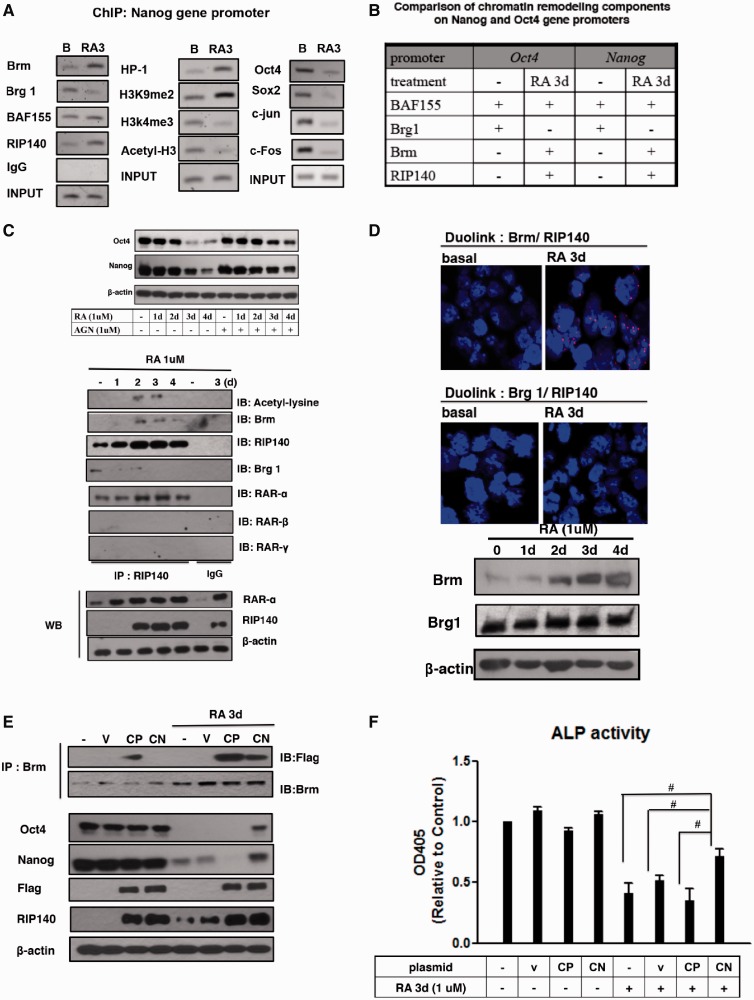
Figure 2.
Acetylated RIP140 mediates the recruitment of Brm complex to repress Nanog and Oct4 gene expression in RA-induced differentiation. (A) ChIP analyses of chromatin-remodeling factors (Brm, Brg1 and BAF155), histone modification markers (H3k4me3, H3k9me2, H3 acetylation and HP1) and transcription factors (c-Fos, c-Jun, Sox2 and Oct4) on Nanog promoter CR1 region. (B) A summary of dynamic changes of relevant chromatin-remodeling components compiled from this current study and the previously published data (7). (C) Top: the effect of RA is mediated by a canonical pathway that is blocked by a pan-RAR antagonist. Bottom: Co-IP analyses to detect lysine acetylation on RIP140 and the interaction between Brg1, Brm and RAR-α. Lower panels show input controls and IgG as a negative control (D) Interaction between RIP140 and Brm, but a lack of interaction between RIP140 and Brg in ESCs. ESCs collected before or after RA stimulation were stained with anti-Brm and -RIP140, or anti-Brg1 and -RIP140, and assessed using the Duolink PLA. Red foci indicate interactions between endogenous Brm and RIP140 proteins. Similar data were observed in ESCs from four different experiments. Brm and Brg1 protein levels after RA treatment were detected by WB. (E) Acetyl-RIP140 associates with Brm in vivo. Co-IP of acetyl mimetic Flag-RIP140 CP (K158/287Q) and acetyl negative Flag-RIP140 CN (K158/287A) with Brm, and the expression of RIP140, Oct4, Nanog and Flag proteins were also detected after RA treatment (1 μM, 72 h). CP, Constitutive positive; CN, constitutive negative; IB, immunoblot. (F) ESCs were treated with control (empty vector), CN or CP, treated with RA 1 for the indicated time intervals. Twenty micrograms of cell lysate was assayed for ALP activity.