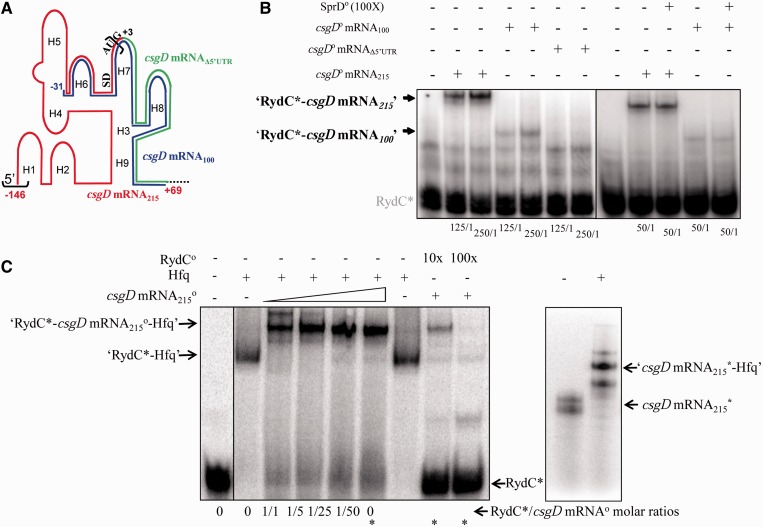
Figure 4.
Direct interaction between RydC and the csgD mRNA; ternary complex formation between RydC Hfq and its mRNA target. (A) Schematic representation of the csgD mRNA 5′-domain emphasizing three RNA constructs. The csgD mRNA215 corresponds to the 215 nt from the 5′-end of the mRNA (red), emphasizing the SD and AUG translation initiation signals. In the csgD mRNA100 variant (blue), 115 nt from the csgD mRNA 5′-end were deleted. The 5′-UTR of the csgD mRNA (the sequence between the black brackets) was deleted in mutant csgD mRNAΔ5′-UTR, therefore starting at G+3 (green). (B) Complex formation between RydC and each of the three csgD mRNA constructs. Native gel retardation assays of purified labelled RydC with increasing amounts of purified unlabelled csgD mRNA215, csgD mRNA100 or csgD mRNAΔ5′-UTR are shown. The csgD mRNA construct/RydC molar ratios are indicated below each lane. Competition assays were performed with a 100-fold molar excess of unrelated purified SprD RNA (18) in the presence of each of the csgD mRNA215 and csgD mRNA100 constructs. (C) Ternary complex formation between RydC, csgD mRNA215 and Hfq. Left panel: Native gel retardation assays show complex formation between labelled RydC and increasing amounts of unlabelled csgD mRNA215 (at a 1- to 50-fold excess as compared with RydC) in the presence or absence of purified Hfq. Hfq is at a 1:1 molar ratio with RydC. The asterisks indicate the ‘RydC*/csgD mRNA’ molar ratio used to perform the competition assays with a 10- to 100-fold molar excess of unlabelled RydC. Right panel: csgD mRNA215 interacts with Hfq in the absence of RydC in vitro. Hfq is at a 20:1 molar ratio with the mRNA. The csgD mRNA adopts two conformations on a native gel.