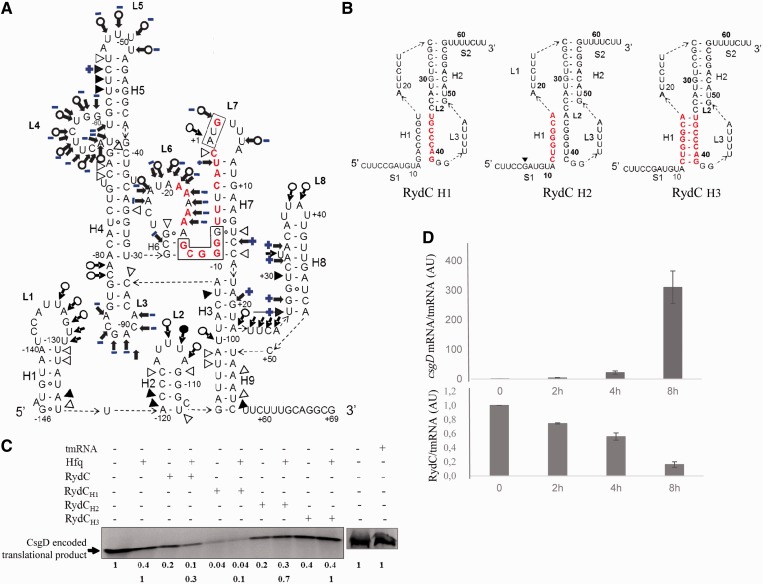
Figure 7.
The interaction between Hfq and csgD mRNA, the role of Hfq in translational regulation and the inverse correlation between RydC and csgD mRNA expression during curli formation in vivo. (A) Secondary structure of csgD mRNA215, with the structural changes induced by Hfq on the csgD mRNA conformation in blue. This model is based on structural probing of the RNA-protein complex in solution (Supplementary Figure S4). The blue plus (+) and minus (−) signs indicate the appearance or disappearance of cleavages induced by the structural probes when the protein is in complex with the mRNA. (B). RydC mutants with mutated nucleotides in red: disrupted stem H1 (RydCH1), stem H1 and the interacting sequence with the csgD mRNA (RydCH2) and a compensatory mutant that restores the H1 structure (RydCH3). (C)In vitro translation of csgD mRNA503 in the presence of various RydC mutants, with and without a 2-fold molar excess of Hfq. The translation products arbitrarily set to 1 were quantified relative to csgD mRNA503 translation in the absence of RydC and Hfq (upper lane). To explore the effect of RydC in the presence of Hfq, the translation products were also set to 1 and quantified relative to CsgD translation in the presence of Hfq (lower lane); tmRNA was used as an internal negative control. (D) qPCR monitoring of csgD mRNA and RydC expression in E. coli cells during curli formation on YESCA plates, normalized against the tmrna reference gene.