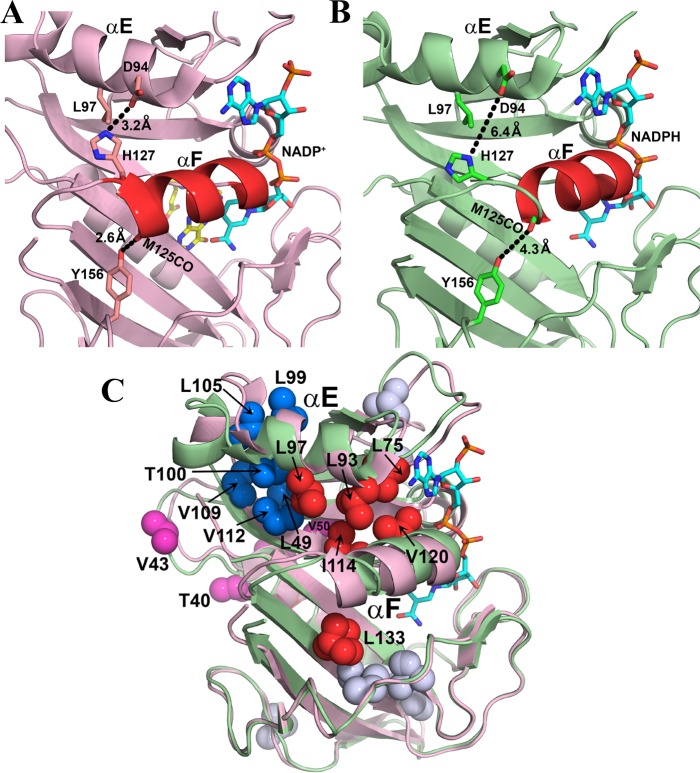
Figure 10.
Role of rotamer averaging in conformational changes associated with hinge motions. (A) Location of helix αF in the X-ray structure of the hE:FOL:NADP+ complex (PDB entry 4M6K).15 Helix αF is stabilized in its hinge-closed conformation by hydrogen bonds, shown by the black dashed lines. (B) X-ray structure of the hE:NADPH complex (PDB entry 4M6J), showing movement of helix αF toward the active site and breaking of the restraining hydrogen bonds upon opening of the hinges.15 Leu97 undergoes increased rotamer averaging in the hE:NADPH complex. Helix αF is colored red in panels A and B, and side chains are shown as sticks. NADP/H is colored cyan and atom colors and folate yellow and atom colors. (C) Superposition of the structures of the hE:NADPH (green) and hE:FOL:NADP+ (pink) complexes, showing conformational changes associated with sliding of helix αF and at the C-terminus of helix αE. Methyl-containing side chains (in PDB entry 4M6J) that undergo rotamer averaging in the hE:NADPH complex are shown as spheres: red for side chains on or in contact with helix αF, blue for side chains that pack against the C-terminal region of helix αE, pink for side chains in hinge 1, and gray for all other methyl-containing side chains with rotamer averaging. Leu133 also exhibits rotamer averaging in the crystal structure of the hE:NADPH complex. The NADPH in PDB entry 4M6J is colored cyan and atom colors; the folate and NADP+ ligands have been omitted from the structure of the hE:FOL:NADP+ complex for the sake of clarity.