Figure 8.
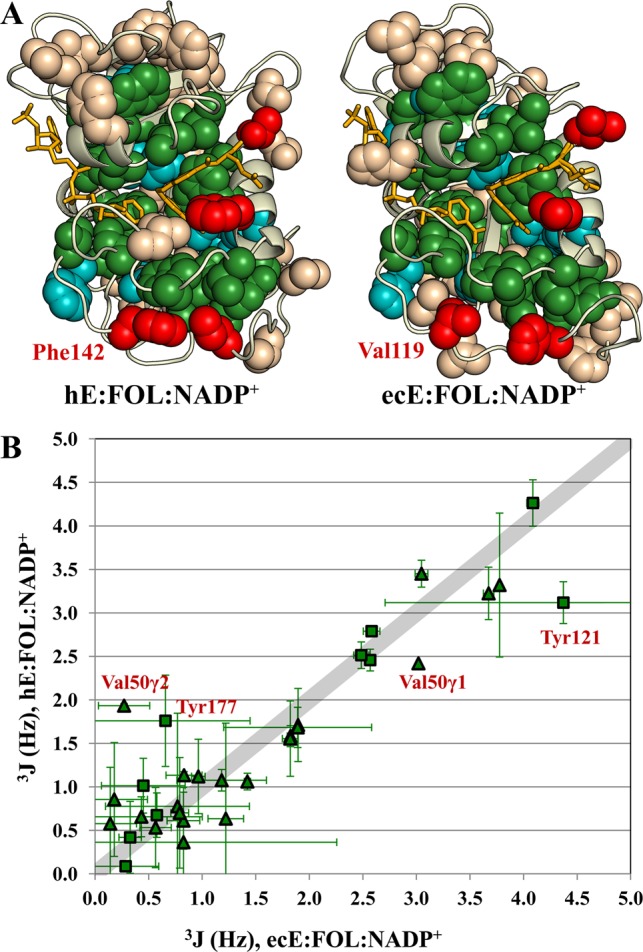
Identical residues in human and E. coli DHFR show remarkably similar average χ1 rotamer conformations. (A) Spheres are shown for Ile, Thr, Val, Leu, and aromatic side chains on the structures of the human and E. coli E:FOL:NADP+ complexes. Identical residues are colored green. Conserved residues (Leu, Val, and Ile in both species or aromatic in both species) are colored cyan. Methyl (aromatic) residues that are aromatic (methyl) residues in the other species are colored red. (B) 3JCγCO and 3JCγN couplings for the identical residues (colored green in panel A) are plotted for the ecE:FOL:NADP+ complex vs the hE:FOL:NADP+ complex. Triangles indicate methyl residues; squares indicate aromatic residues. Val50 and Tyr121, which deviate the most from the line with a slope of 1 (broad gray line), are sensitive to the bound ligands in both human and E. coli DHFR.
