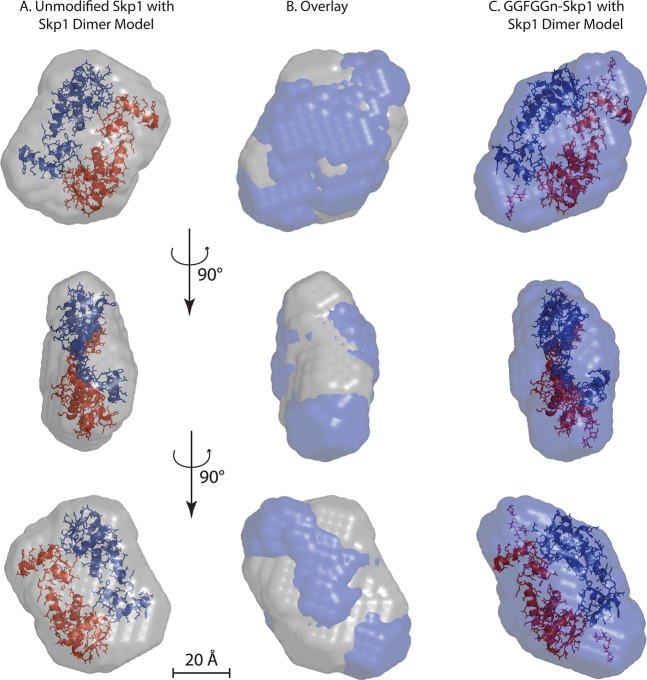
Figure 5.
SAXS envelopes. Ab initio molecular envelopes were calculated from data collected from 9.4 mg/mL unmodified Skp1 (A) or 3.0 mg/mL GGFGGn-Skp1 (C), based on the average of 10 iterations using DAMMIF. (B) Overlay of panels A and C. In panels A and C, images of two Skp1 molecules excerpted from the crystal structure of a complex with the FBP Tir1 (Protein Data Bank entry 2P1N)46 are modeled with P2 symmetry within the envelope of Skp1 and GGFGGn-Skp1. The Skp1 pentasaccharide was modeled using GLYCAM, and the reducing terminus is positioned near the Pro143 attachment site. The bottom two rows represent 90° relative rotations as indicated.