Abstract
Successful innovative proteomic analysis is highly dependent on molecular biology techniques at the surveying and validation stage. This is because mass spectrometry (MS) analyses of complex samples are limited by their dynamic range for detection—so careful front-end sample preparation, fractionation, and enrichment are crucial to find biologically relevant signals in an extremely complex extracellular environment. Here, we share a very useful and simple front-end surveying methodology—lectin blotting—for proteomic analysis of glycosylation patterns—the most abundant posttranslational modification in extra-cellular signaling.
Lectin blotting is an effective biochemical technique based on lectin–glycan interactions. It is used to detect and characterize carbohydrate epitopes on protein or lipids. Depending on lectin patterns, specific lectins can be used as tags to enrich glycoproteins for further proteomic analysis. In this method, proteins or lipids are resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred electrophoretically to adsorbent membranes such as nitrocellulose or polyvinylidene difluoride (PVDF) membrane first. Transferred proteins or lipids are subsequently analyzed by probes including labeled lectins. In this chapter, we provide a detailed methodology of lectin blotting for glycol-proteomic analysis. This method is robust and can be used for complex cell lysates, conditioned media, or human samples to monitor glycosylation pattern following extracellular signal transduction. Here, we will use patient plasma samples post hypothermic therapy—an extremely complex medium—as a tool to describe in detail the technique of lectin blotting for proteomic analysis.
Keywords: Lectin blot, Glycoprotein, Glycosylation, Glycoproteomics
1 Introduction
Glycosylation is a common and important form of posttranslational modification. It has been reported that more than 50 % of human proteins are likely conjugated with glycans [1]. Glycosylation, especially N-linked glycosylation, is prevalent in proteins destined for extracellular environments, including clinically accessible fluid—plasma. Glycosylation plays vital roles in a variety of biologic processes such as cell–cell communication, cell matrix interaction, adhesion, and protein targeting. Changes of glycosylation status are usually highly related to pathophysiological changes. Thus, the diagnostic and therapeutic potentials in decoding protein glycosylation patterns have been recognized [2, 3] and we feel this process is worthy of investigation in the extracellular milieu.
Recent advances in MS/glycomic technology enables in-depth characterization that utilizes differential lectin binding to broadly characterize complex media such as cell lysates, conditioned media, and human plasma to study in cell–cell signaling. However MS analyses of complex samples are limited by their dynamic range for detection—so careful front-end sample preparation, fractionation, and enrichment are key steps to find biologically relevant signals in an extremely complex extracellular environment. Before in-depth characterization, it is recommended that glycosylation pattern changes with respect to treatment state should be profiled because glycosylation patterns are like “zip codes”—so instead of searching on random streets to pull out just long lists of proteins, we can focus on “neighborhoods” of interest and screen for areas that changed the most due to treatment or disease state. Lectin blotting is an efficient screening method for profiling protein glycosylation status.
Lectins are carbohydrate-binding proteins that may interact with high specificity to a soluble carbohydrate or to a carbohydrate moiety that is a part of a glycoprotein or a glycolipid. Since the first lectin was discovered more than 100 years ago in plants, lectins have been found to occur ubiquitously in nature and have been widely used as an effective biochemical tool [4, 5]. For example, concanavalin A and other commercially available lectins have been commonly used in affinity chromatography for purifying glycoproteins, in lectin arrays and lectin blotting for detecting carbohydrate moieties [6–8]. Here, we discuss a lectin blotting method in detail.
2 Materials
Prepare all solutions using ultrapure water and analytical grade reagents. Prepare and store all reagents at room temperature (unless indicated otherwise). Diligently follow all waste disposal regulations when disposing waste materials. We do not add sodium azide to the reagents.
2.1 Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis Components
SDS lysis buffer (2×): Premix Laemmli Sample Buffer (Bio-Rad) with an equal volume of protein sample buffer containing 62.5 mM Tris–HCl (pH 6.8), 25 % glycerol, 2 % SDS, and 0.01 % bromophenol blue in ultrapure water.
SDS-polyacrylamide gel: Precast gels from Life Technologies. Novex® 4–20 % Tris-Glycine Mini Gel (1.0 mm, 12 wells) are polyacrylamide gels based on traditional Laemmli protein electrophoresis.
SDS-PAGE system from Invitrogen.
SDS-PAGE running buffer: Final concentration of 25 mM Tris, 192 mM glycine, 0.1 % SDS in ultrapure water.
Human plasma samples and astrocyte cell culture-conditioned medium (DMEM for control and oxygen glucose deprivation (OGD)/rescue; glucose-free DMEM medium for OGD) concentrated by ultrafiltration centrifugation with 10 kDa cutoff membrane, according to the manufacturer’s instructions.
2.2 Lectin-Blotting Components
Protein blotting transfer system: iBlot® 7-Minute Blotting System bought from Life Technologies is used to blot proteins from gels to nitrocellulose or PVDF membranes. The iBlot® 7-Minute Blotting System can blot proteins from polyacrylamide gels in about 7 min without the need for additional buffers or an external power supply using disposable blotting stacks with integrated nitrocellulose or PVDF membranes.
Tris-buffered saline (TBS; 10×): Final concentration: 1.5 M NaCl, 0.1 M Tris–HCl, pH 7.4, in ultrapure water.
Tris-buffered saline-Tween (TBST): TBS containing 0.05 % Tween-20. To make 1 L TBST, add 100 mL of 10× TBS to a 1 L graduated cylinder and adjust volume to 1 L with deionized water. Add 500 μL of Tween-20 (see Note 1).
Blocking solution: 3 % bovine serum albumin (BSA) in TBST. Recommend solution is freshly prepared prior to use. If stored at 4 °C, use within 1 week.
Lectin Kit 1 (Table 1, including eight kinds of biotinylated lectins) and streptavidin–horseradish peroxidase [HRP] conjugate (Vector lab), Pierce enhanced chemiluminescence (ECL) Western Blotting Substrate, Thermo Scientific CL-XPosure Film, Kodak, M35A-M X-OMAT Processor for imaging and autoradiograph film/cassette.
Table 1.
Lectins and their primary sugar specificity
| Lectin | Common abbreviation | Primary sugar specificity |
|---|---|---|
| Concanavalin A | Con A | Mannose |
| Dolichos biflorus agglutinin | DBA | N-Acetylgalactosamine |
| Ricinus communis agglutinin 1 | RCA 1 | Galactose, N-Acetylgalactosamine |
| Soybean agglutinin | SBA | N-Acetylgalactosamine |
| Ulex europaeus agglutinin 1 | UEA 1 | Fucose |
| Wheat germ agglutinin | WGA | N-Acetylglucosamine |
| Maackia amurensis lectin I | MAL I | Galactosyl (β-1,4) N-acetylglucosamine structures |
| Maackia amurensis lectin II | MAL II | Sialic acid in an (α-2,3) linkage |
3 Methods
All the procedures should be performed at room temperature unless otherwise specified.
Dilute the plasma samples quantified by Bradford method to 2 μg/μL with deionized water and add SDS lysis buffer (2×) at 1 μg/μL final concentration (see Note 2). For cell culture medium, concentrated by ultrafiltration centrifugation with 10 kDa cutoff membrane to 15 μL in 30 min, protein concentration is also determined by Bradford method.
Heat samples at 95 °C for 5 min to denature proteins.
Centrifuge the heated samples at 12,000 × g for 5 min to bring down the condensate (see Note 3).
Put Novex® 4–20 % Tris-Glycine Mini Gel into electrophoresis box (see Note 4).
Load samples into the wells (see Note 5).
Run the gel at a constant voltage of 120 V.
Stop running the gel when the bromophenol blue front reaches the bottom of the gel (see Note 6).
Rinse the gel in deionized water to remove traces of SDS-PAGE running buffer.
To prepare for the trans-blot, open the package of iBlot® Transfer Stack, PVDF mini.
Place the gel on the PVDF membrane in the “sandwich” chamber (see Note 7).
Run the trans-blot at room temperature for 7 min (see Note 8).
After the trans-blot, block the membrane (facing up) with blocking solution for at least 1 h at room temperature or 4 °C overnight, on the shaker (see Notes 9 and 10).
Incubate with a 3 μg/mL biotinylated lectin solution in blocking buffer on a shaker for 30 min (see Note 11).
Wash the membrane with TBST on a shaker: twice for 5 min, and then twice for 10 min.
Incubate with 0.1 μg/mL streptavidin–HRP conjugate in blocking buffer for 20 min (see Note 12).
Rinse the membrane with TBST on a shaker: twice for 5 min, and then thrice for 10 min.
Prepare one tube for Pierce ECL Western Blotting Substrate solution (3 mL for each gel).
Put the PVDF membrane on a smooth, clean surface, such as a glass plate or a parafilm. Pour substrate solution on the membrane and incubate in the dark for about 2 min (see Note 13).
After incubation, absorb any excess liquid.
Proceed to the darkroom with the following items: timer, and autoradiograph film/cassette.
Put a film on the incubated PVDF membrane to develop. Developing times can vary depending on the relative amounts of glycosylation present.
Feed membrane, film, and cassette into a preheated film developer (the machine should be switched on 30 min before film development). Wait for the film to demonstrate distinct signals (Figs. 1 and 2).
Fig. 1.
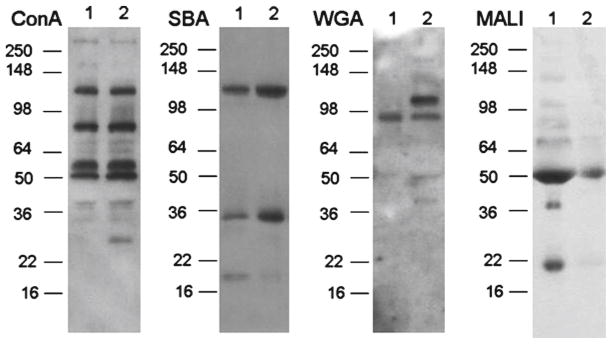
Examples of lectin blot surveys conducted in plasma. Lane 1 plasma sample post hypothermic therapy with poor clinical outcome; Lane 2 plasma samples post hypothermic therapy with good clinical outcome. SDS-PAGE molecular weight standards are marked on the left of each film
Fig. 2.
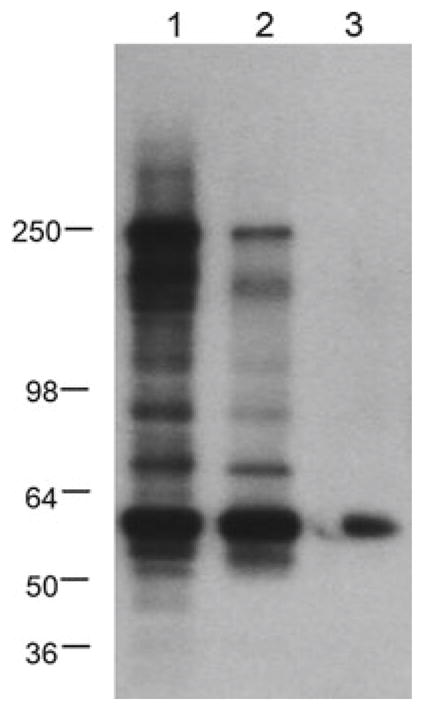
Example of lectin blot surveys conducted in conditioned cell culture medium and lectin blot with ConA. Lane 1 control, Lane 2 post-oxygen glucose deprivation (OGD), Lane 3 post-OGD/rescue. SDS-PAGE molecular weight standards are marked on the left
Acknowledgments
This work was supported by National Institutes of Health/National Institute of Neurological Disorders and Stroke grants R01 NS067139 (to MMN) and P01 NS55104 (to EHL).
Footnotes
Cutting the end of a blue 200–1,000 μL pipette tip facilitates aspirating Tween-20 easily because Tween-20 is viscous liquid.
SDS precipitates at 4 °C. Therefore, the lysis buffer needs to be warmed prior to use.
Centrifuging the samples prior to the run helps remove insoluble debris.
Gel gradient percentage is dependent on your requirement.
Samples containing multiple proteins require 1–20 μg of protein in 2–10 μL per well. Purified samples containing single or very few proteins require less (0.5–5 μg in 2–10 μL).
For certain targeted proteins, it is not necessary to run dye to the bottom. Run SDS-PAGE as appropriate for the protein of interest.
Hold the two top corners of the gel with each hand. Lower the bottom part of the gel first on the lower part of the membrane and gently release the gel little by little to lay the complete gel on the membrane. This will prevent trapping of bubbles in between the gel and the membrane. A blotting roller is used to remove any air bubbles between the gel and blotting membrane during the assembly of the stacks and gel.
Transfer time can be varied according to the targeted protein’s molecular weight. Like in any electrophoresis separation, small proteins (under 30 kDa) migrate more rapidly than large ones and, in the same manner, need less time to transfer from the gel’s matrix to the membrane. Seven minutes in this system works well with most proteins; less time is needed to transfer smaller proteins with the iBlot (i.e., 5–6 min). Larger proteins (over 150 kDa) migrate slower than smaller proteins during SDS-PAGE and therefore need more transfer time. The recommended time to transfer large proteins with the iBlot is 8–10 min.
In Western blotting, nonfat dry milk is usually used as blocking reagent. However, in lectin blotting, BSA is used as blocking reagent because it is not a glycoprotein. Blocking reagents that contain even small amounts of glycoproteins (e.g., nonfat dry milk) may bind to the lectins, potentially depleting the available lectin or producing a high background signal. Alternatively, gelatin can be used as a blocking reagent [9].
The used gel should be disposed of as hazardous waste.
The concentration of biotinylated lectin and incubation time can be regulated according to the type of lectin. Depending on the lectin’s carbohydrate specificity, it may be necessary to optimize the working dilution.
The concentration of streptavidin–HRP conjugate and incubation time should be optimized according to the type of lectin.
Substrate solution should be made fresh. A membrane for mini gel needs 2 mL substrate solution.
References
- 1.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 2.Dube DH, Bertozzi CR. Glycans in cancer and inflammation potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 3.Cao J, Shen CP, Wang H, Shen HL, Chen YH, Nie AY, et al. Identification of N-glycosylation sites on secreted proteins of human hepatocellular carcinoma cells with a complementary proteomics approach. J Proteome Res. 2009;8:662–672. doi: 10.1021/pr800826u. [DOI] [PubMed] [Google Scholar]
- 4.Sharon N, Lis H. History of lectins: from hemagglutinins to biological recognition molecules. Glycobiology. 2004;14:53R–63R. doi: 10.1093/glycob/cwh122. [DOI] [PubMed] [Google Scholar]
- 5.Sharon N. Lectins: past, present and future. Biochem Soc Trans. 2008;36:1457–1460. doi: 10.1042/BST0361457. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Hancock WS. Monitoring glycosylation pattern changes of glycoproteins using multi-lectin affinity chromatography. J Chromatogr A. 2005;1070:57–64. doi: 10.1016/j.chroma.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 7.Balcan E, Gümüş A, şahin M. The glycosylation status of murin postnatal thymus: a study by histochemistry and lectin blotting. J Mol Histol. 2008;39:417–426. doi: 10.1007/s10735-008-9180-3. [DOI] [PubMed] [Google Scholar]
- 8.Uchiyama N, Kuno A, Koseki-Kuno S, Ebe Y, Horio K, Yamada M, Hirabayashi J. Development of a lectin microarray based on an evanescent-field fluorescence principle. Methods Enzymol. 2006;415:341–351. doi: 10.1016/S0076-6879(06)15021-1. [DOI] [PubMed] [Google Scholar]
- 9.Freeze HH. Lectin analysis of proteins blotted onto filters. Curr Protoc Mol Biol. 2001;Chapter 17(1993) doi: 10.1002/0471142727.mb1707s23. Pages: Unit 17.7. [DOI] [PubMed] [Google Scholar]


