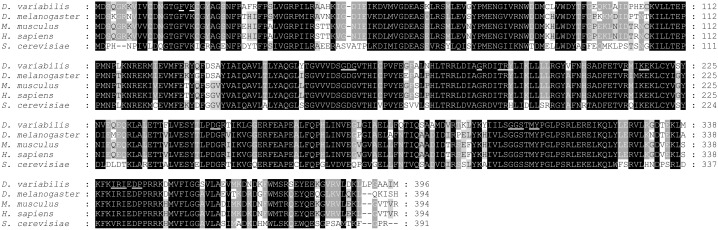
Figure 1. Tick Arp2 subunit multiple sequence alignment and identification of conserved ATP binding sites.
Multiple sequence comparison by log-expectation (MUSCLE) software was used to create a sequence alignment of Arp2 subunits from D. variabilis, D. melanogaster, M. musculus, H. sapiens, and S. cerevisiae. Identical and similar amino acids are highlighted in black and grey, respectively. Conserved ATP binding sites predicted by the NsitePred web server are underlined.