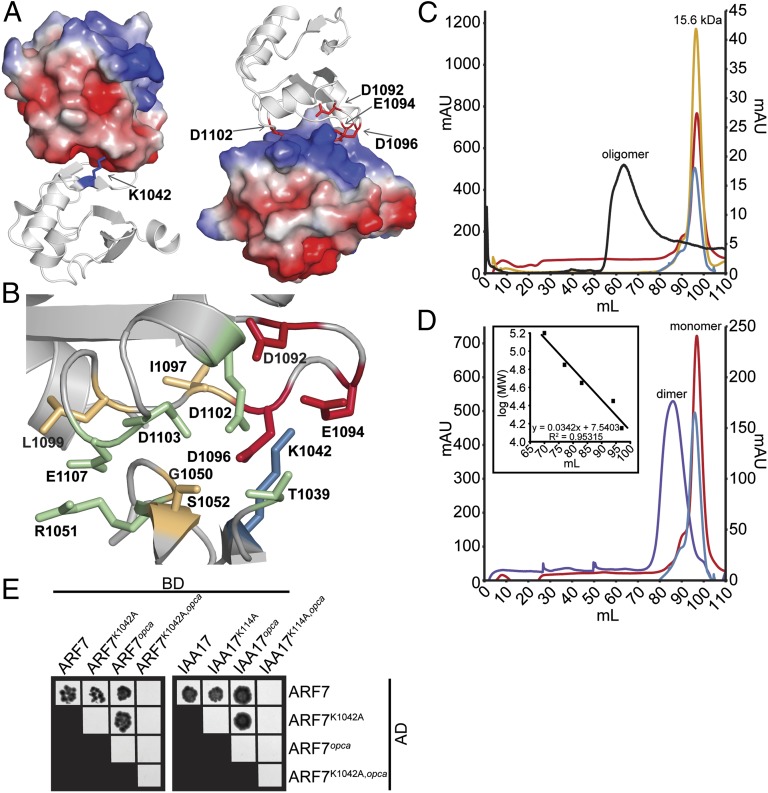
Fig. 2.
ARF and Aux/IAA PB1 domain interactions are driven by charge–charge interactions. (A) The modeled ARF7PB1 electrostatic surface potential reveals positive (blue) and negative (red) interaction interface, containing the invariant lysine and OPCA motif, respectively. (B) PISA predictions of residue contributions to interactions identified the ARF7PB1 interaction interface containing the invariant lysine (blue), OPCA motif (red), and residues important for hydrogen bonding (orange) and charge–charge interactions (green). (C) Size-exclusion chromatography reveals that wild-type ARF7Cterm exists as soluble aggregate in solution (black), whereas mutations in the invariant lysine (blue), the OPCA motif (red), or both (yellow) afford monomeric protein. (D) Size-exclusion chromatography reveals that mixing ARF7CtermK1042A (blue) and ARF7Ctermopca (red), which exist as monomeric protein in solution, results in the formation of a dimer (ARF7CtermK1042A and ARF7Ctermopca mixed in a 1:1 ratio, purple). Inset, protein molecular weight standards. (E) Yeast two-hybrid assays reveal that introduction of single unlike PB1 mutations in ARF7 and IAA17 does not affect ARF–ARF or ARF–Aux/IAA interactions. Combining both PB1 mutations (lysine and opca) or like mutations—e.g., both proteins contain lysine or OPCA mutations—abrogates protein–protein interactions. BD, binding domain; AD, activation domain.