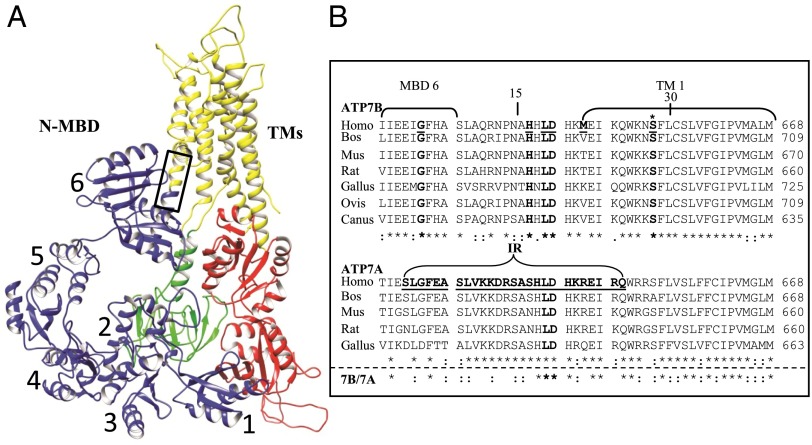
Fig. 1.
Hypothetical ATP7B model and multiple species alignment of the conserved regions, amino acids 621–668, in the two Cu-ATPases. (A) A hypothetical ATP7B ribbon model, generated by UCSF Chimera, showing the conserved core organization (20). The two large cytoplasmic loops in the core structure are: the A domain (actuator, green), between TM4 and TM5, which contains the phosphatase activity; and the N and P domains (nucleotide binding and phosphorylation, red) between TM6 and TM7, which bind ATP (N), catalyzing formation of a phosphorylated intermediate (P) as part of the catalytic cycle. The eight TMs (yellow are): TM1 (amino acids 645–670), TM2 (including the platform helix, amino acids 694–722), TM3 (amino acids 729–749), TM4 (amino acids 765–786), TM5 (amino acids 916–942), TM6 (amino acids 967–1004), TM7 (amino acids 1307–1345), and TM8 (amino acids 1352–1373). The six N-terminal MBDs (blue, N-MBDs, also referred to as the N-terminal domain of ATP7B, N-ATP7B) (61, 62) were manually positioned onto the published model. The box approximates the region of the multiple species alignment shown in B. (B) A multiple species alignment of human ATP7B sequence 621–668 (Upper) and ATP7A sequence 621–668 (Lower). WD patient mutations are underlined and in bold. The ATP7B S653 position is marked with an asterisk (bold). Portions of MBD6 and TMD 1 are bracketed. In ATP7A, the bracketed sequence shows the deleted region that is replaced with two amino acids (IR) in a patient with Occipital Horn Syndrome. The deleted sequence of ATP7A is underlined and in bold (35). Alignments were obtained using ClustalW (63). Amino acids that are identical (*), conserved (:), and semiconserved (.) are shown.