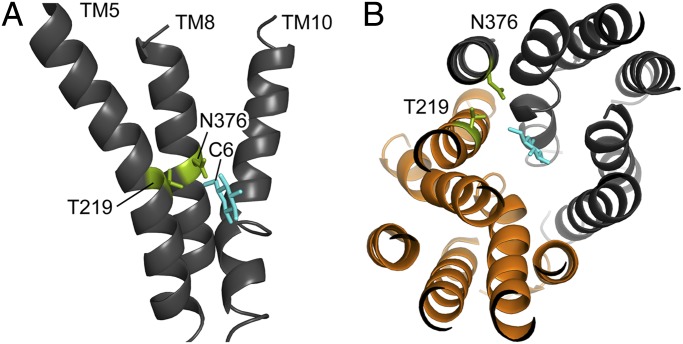
Fig. 3.
Homology model of the Gal2 structure. The model is based on the outward-facing partly occluded structure of E. coli XylE with bound glucose (PDB ID code 4GBZ). (A) Side view of Gal2; for reasons of clarity, only TMs 5, 8, and 10 are shown. The two amino acid residues T219 and N376 (green) are located at the center of their respective helix, with their side chains protruding toward the C6 of glucose (cyan). (B) Top view of Gal2 from the extracellular side, with a cross-sectional plane for better view; glucose (cyan) is found in between subdomains N (orange) and C (dark gray). The 3D images were created with PyMOL (41).