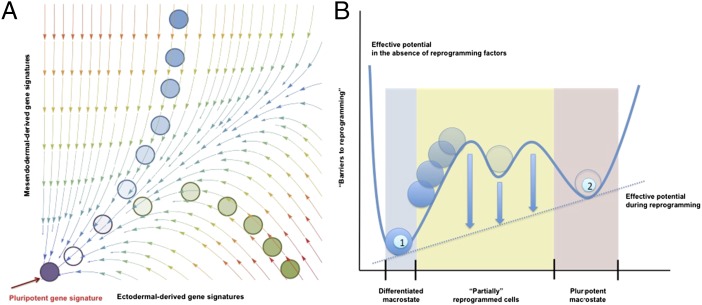
Fig. 1.
Comparison of mathematical models derived from single-cell and population-level analyses. (A) Schematic representation of a probability landscape as determined from gene network analysis at the single-cell level. The planar axes represent gene expression levels for two different genes G1 and G2; a vertical component would represent the probability of a given cell to express the defining genes (G1 and G2) at a specific level. The arrows represent the curl flux forces, and the colors represent the magnitudes of the potential. The motion of a cell in this space is governed by both the curl flux and the shape of the potential landscape. For simplification, reprogramming has been depicted as the changes in gene expression from mesendodermal- and ectodermal-derived somatic lineages (e.g., cardiomyocytes and hepatocytes serve as a representative mesendodermal-derived lineages, whereas neurons would represent and ectodermal origin). (B) Effective potential landscape determined from population-level analyses. This potential landscape reduces many of the complexities at the single-cell level into a 1D effective potential energy function in cell state space. Higher potential energies are represented by larger heights. The numbers refer to the differentiated somatic cell state (1) and the pluripotent state (2). Note that population approaches consider variable macrostates in which different individual gene signatures are equally represented.