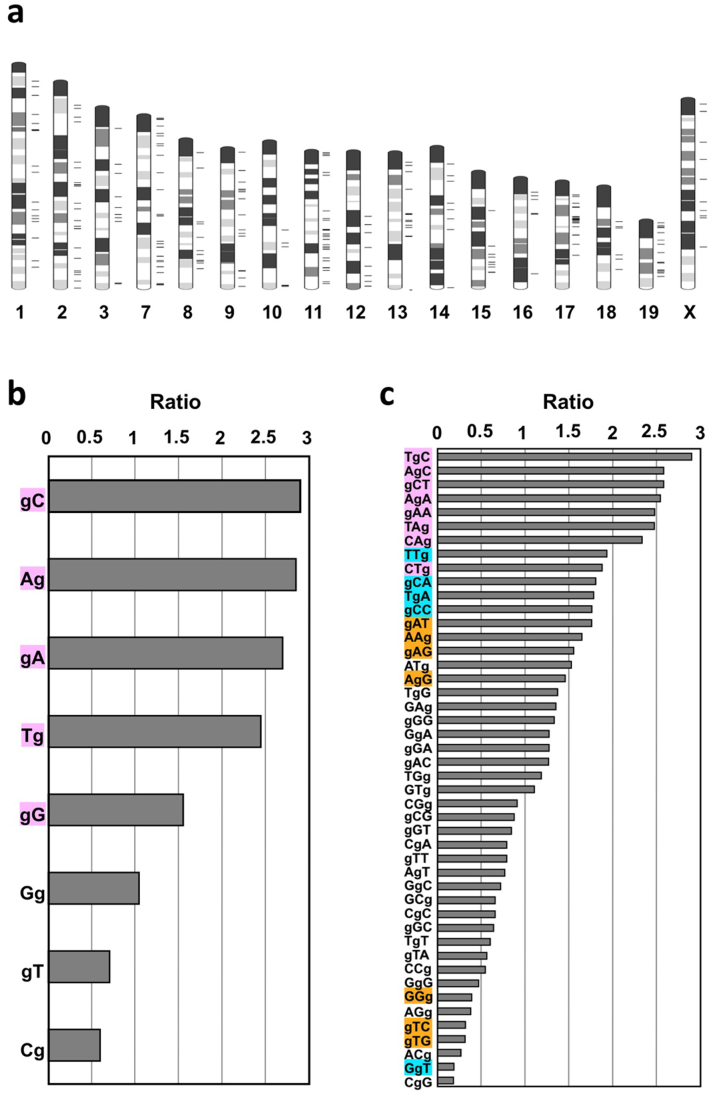
Figure 5. Genome-wide distribution of mutations and site preferences of G to T mutations in di- and trinucleotide sequences.
(a) Mutations detected in G2–G8 were mapped on a mouse G-band ideogram using Ideographica (http://www.ncrna.org/idiographica/). Each black transverse line on the right side of the chromosome represents a mutation site. (b) Site preferences of G to T mutations in di-nucleotide sequences. The plots represent the relative ratio of the actual value of detected mutations (G to T mutations in G2–G8) in each di-nucleotide to its occurrence level in the analyzed exome sequences. ‘g' indicates the position of a mutated guanine. (c) Site preferences of G to T mutations in tri-nucleotides. For each nucleotide sequence, a chi square test (detected vs. expected) was performed, and the colored sequences indicate a significant difference: P < 0.001 (pink), P < 0.01 (blue), and P < 0.05 (orange).