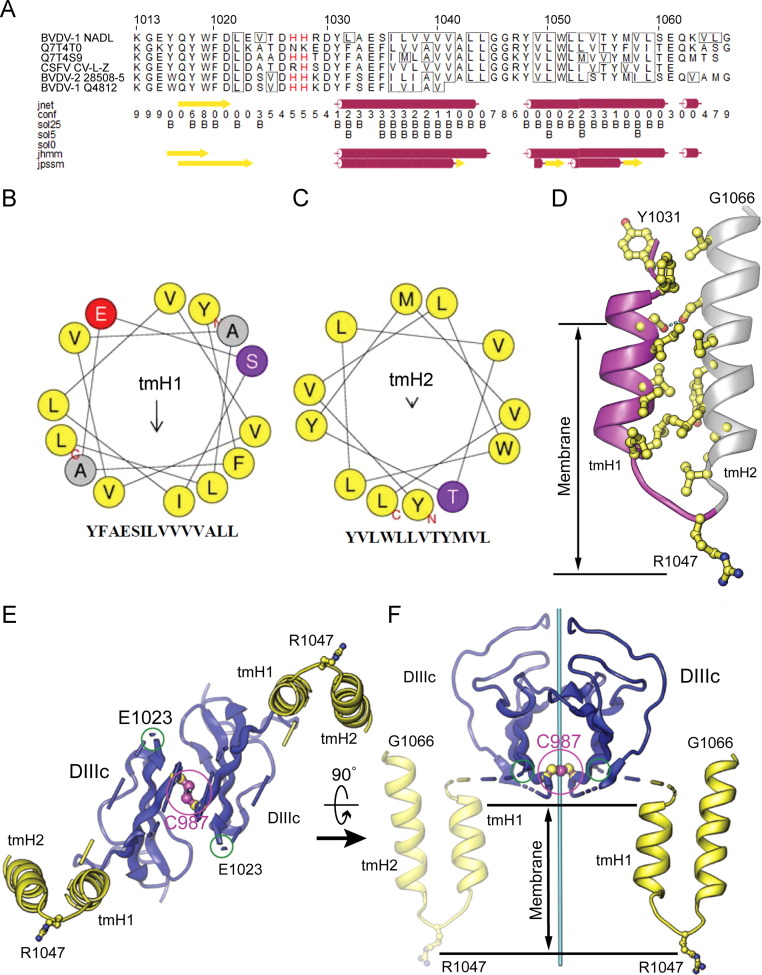
Fig. 2.
Sequence analysis and atomic modeling of the BVDV E2 membrane anchor. (A) Two transmembrane helices (tmH1 and tmH2) were predicted with JPRED-3. The reference sequence and numbering refer to the BVDV-1 NADL polyprotein (Xu et al., 1997). This sequence was used to query the UniRef90 database. The following pestivirus E1 UniProt sequences returned from the search are shown: Q7T4T0, pestivirus PG-1; Q7T4S9, border disease virus strain V2536/2; Q9WPE6, classical swine fever virus strain C-V-LZ; Q9WP29, BVDV-2 strain 28508-5; Q65793, BVDV-1 strain Q4812. (B and C) Helical wheel representations of the core portions of tmH1 (B), and tmH2 (C). The arrows represent the direction and magnitude of the amphipathic moment of each helix. (D) Computationally generated model of the E2 membrane anchor. An interhelical hydrogen bond (dashed line) stabilizes the otherwise hydrophobic packing between the two helices of the hairpin. (E and F) Two orthogonal views of a computational model for dimeric E2 domain IIIc (DIIIc) followed by the membrane anchors. The positions and orientations of the membrane anchors relative to the DIIIc domains may vary depending on the structure of the linker between DIIIc and the anchor (blue/yellow dashed lines). C987 and the last residue in the crystal structure, E1023 are circled in magenta and green, respectively. R1047, which stabilizes the short helical hairpin by “snorkeling” to the inner surface of the viral membrane, is in ball-and-stick representation. The E2–E2 dyad is shown in cyan.