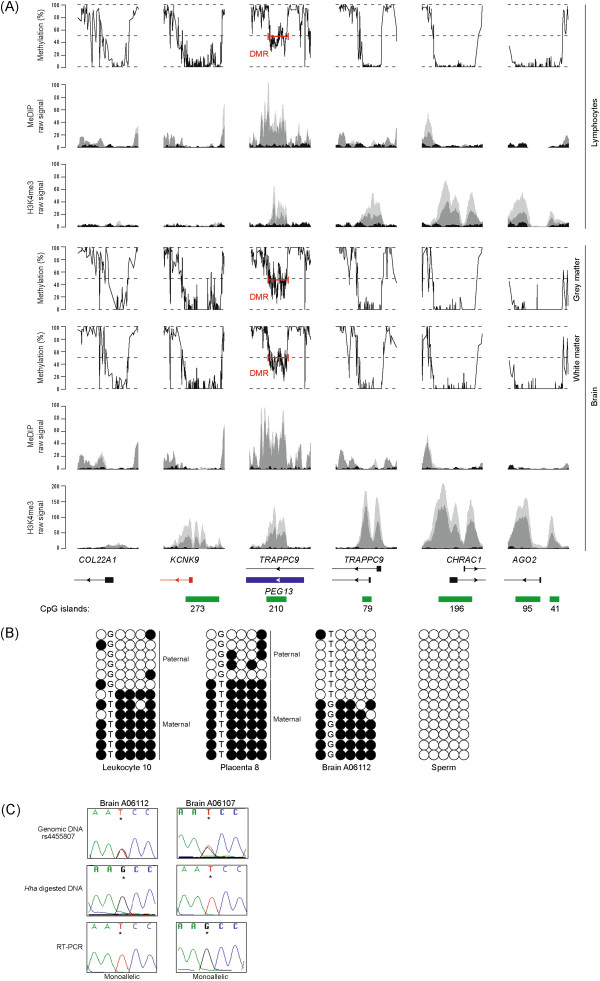
Figure 2.
DNA methylation profiling of gene promoter of genes flanking TRAPPC9/PEG13. (A) CpG methylation profiling in lymphocytes and brain samples as defined by WGBS. The analyses were restricted to the intervals overlapping the promoters/CpG islands and are associated with the expected H3K4me3 and meDIP signatures in CD4 lymphocytes cells and fetal brain. The two shades of grey peaks in the meDIP and ChIP-seq panels represent two independent biological replicates compared to input (black peaks) with the y-axis showing the number of ChIP-seq reads. The precise location of the PEG13-DMR, as defined by approximately 50% methylation and co-enrichment of H3K4me3 and meDIP is indicated by the red bracket. (B) The methylation status of the PEG13-DMR was confirmed using standard bisulphite PCR on DNA samples heterozygous for SNP rs4455807. (C) Sequence traces for two brain samples revealing the genotypes of the PEG13-DMR methylated allele (resistant to HhaI digestion) and expression from the unmethylated allele.