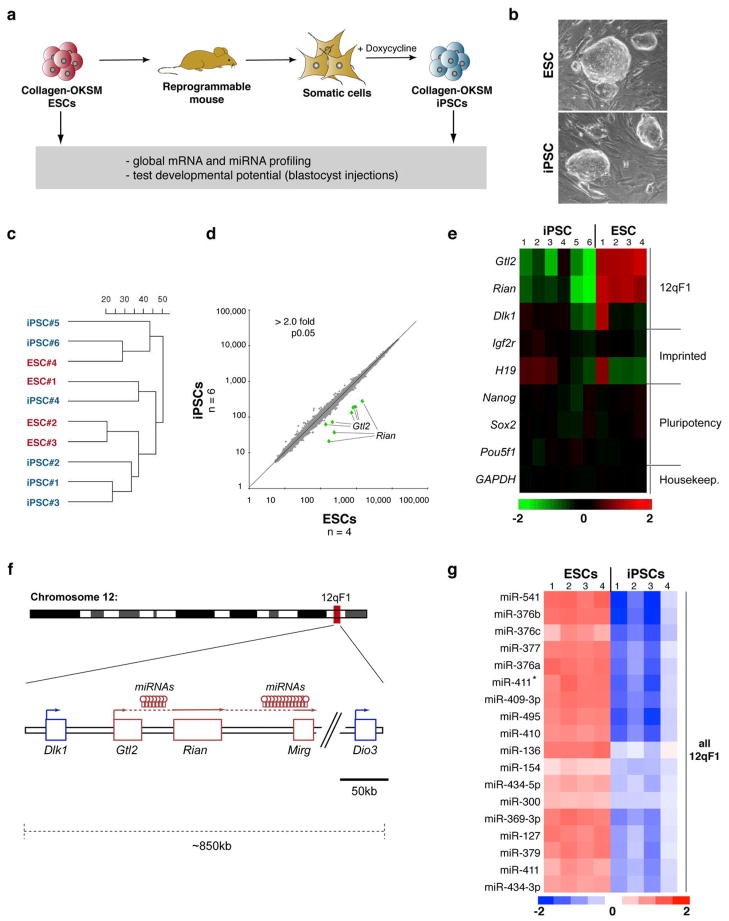
Figure 1. Aberrant silencing of the Dlk1-Dio3 gene cluster in mouse iPSCs.
(a) Strategy for comparing genetically matched ESCs and iPSCs using “reprogrammable mice” harboring a doxycycline-inducible polycistronic reprogramming cassette (OKSM) in the Col1a1 (Collagen) locus. (b) Morphology of Collagen-OKSM ESCs and iPSCs. (c) Unsupervised clustering of four ESC and six iPSC lines based on microarray expression data. (d) Scatterplot of microarray data comparing iPSCs and ESCs with differentially expressed genes highlighted in green (2-fold, p0.05, t-test with Benjamini-Hochberg correction). (e) Heatmap showing relative expression levels of selected mRNAs in ESCs and iPSCs, covering in addition to Gtl2 and Rian other imprinted genes (Dlk1, Igf2r and H19) and pluripotency-associated transcripts (Nanog, Sox2 and Pou5f1). (f) Schematic representation of mouse chromosome 12 with position of the Dlk1-Dio3 gene cluster highlighted. Maternally-expressed and paternally-expressed transcripts are shown in red and blue, respectively. (g) Heatmap showing miRNAs that are differentially expressed between ESCs and iPSCs (2-fold, p0.01, t-test).