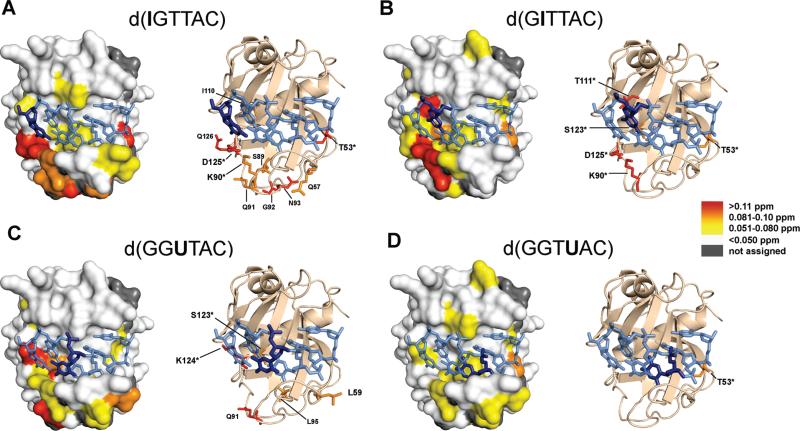
Figure 7.
(Left) Surface representation of the high-resolution crystal structure of the Pot1pN/d(GGTTAC) complex (27) highlighting the MCSPs resulting from the binding of each substituted oligonucleotide, G1I, G2I, T3dU, and T4dU (site of variation highlighted in dark blue on a stick model of DNA). (Right) Cartoon/ribbon model of the Pot1pN/d(GGTTAC) complex highlighting the side chains of specific residues undergoing moderate and strong perturbations in each Pot1pN/substituted nucleotide complex. Residues that make direct contact with the oligonucleotide in the Pot1pN/d(GGTTAC) complex are designated with an asterisk. (A) Moderate and strongly perturbed residues in Pot1pN/d(IGTTAC) complex occur on loop1–2 (T53* and Q57), loop3–4 (S89, K90*, G92, and N93), β4 (I110), and β5 (D125* and Q126). (B) Moderate and strongly perturbed residues in the Pot1pN/d(GITTAC) complex occur in loop1–2 (T53*), loop3–4 (K90*), β4 (I111*), and β5 (S123* and D125*). (C) Moderate and strongly perturbed residues in the Pot1p/d(GGUTAC) complex occur in loop1–2 (L59), loop3–4 (Q91 and L95), and β5 (S123* and K124*). (D) The only moderately perturbed residue in the Pot1pN/d(GGTUAC) complex is found on loop1–2 (T53*).