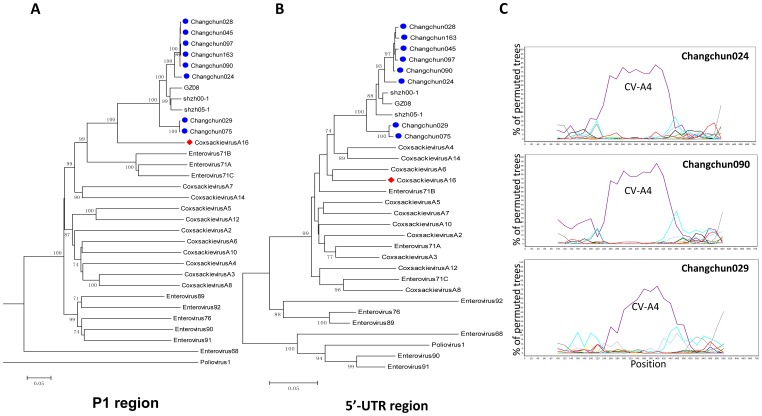
Figure 2. Phylogenetic analysis of the 5′UTR and P1 regions of eight circulating Changchun CV-A16 strains.
(A) The neighbor-joining tree was generated based on the 5′UTR sequences (nucleotide 24–718 using CV-A16-G10 sequence as reference) of eight circulating CV-A16 viruses and the HEV reference sequences. (B) The neighbor-joining tree was generated based on the P1 sequences (nucleotides 751–3327 using the CV-A16-G10 sequence as the reference) of eight circulating CV-A16 viruses and reference sequences. The ▪icon indicates CV-A16 strains isolated from Changchun; ♦icon indicates the prototype CV-A16-G10.(C) Detailed bootscanning analysis of the 5′UTR region from CV-A16 strains circulating in Changchun. Bootscanning analysis was performed based on the 5′UTR region sequences (nucleotides 2–718 using the CV-A16-G10 sequence as reference) of Changchun024 and Changchun029, using HEV type A sequences and poliovirus 1 and EV68 as outgroups. A sliding window size of 200 bases and step size of 20 bases was used.