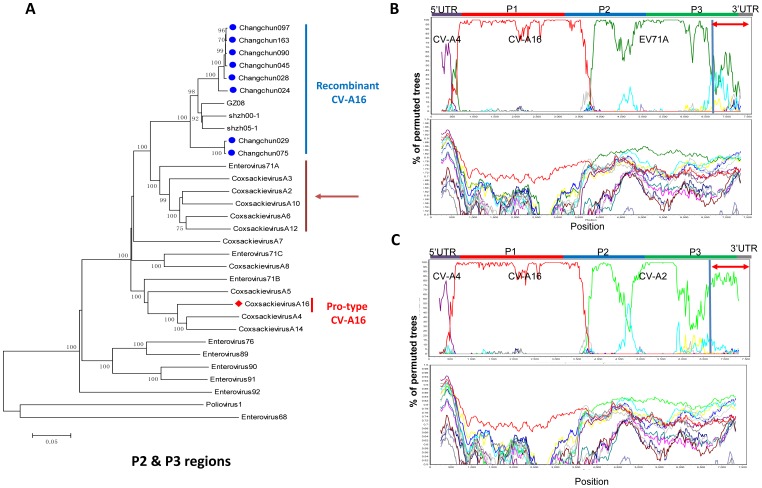
Figure 3. Phylogenetic analysis of P2 and P3 regions of eight circulating Changchun CV-A16 strains.
(A) Phylogenetic analysis of the P2 and P3 sequences of circulating CV-A16 stains (nucleotides 3341–7328 using the CV-A16-G10 sequence as the reference). Only strong bootstrap values (>70%) are shown.▪icon indicates CV-A16 strains isolated from Changchun; ♦icon indicate the prototype CV-A16-G10. (B) Bootscanning analysis of Changchun024 complete genomic sequences was performed with all HEVA sequences except CV-A2, CV-A3, CV-A6, CV-A10 and CV-A12. (C) Bootscanning analysis of Changchun024 was performed with all HEVA sequences except EV71,CV-A3, CV-A6, CV-A10 and CV-A12. (D) Bootscanning analysis of Changchun024 was performed with all HEVA sequences except EV71, CV-A2, CV-A3, CV-A6 and CV-A10. Poliovirus 1 and enterovirus 68 were used as outgroups. A window size of 500 bp and step size of 20 bp at a time were used.