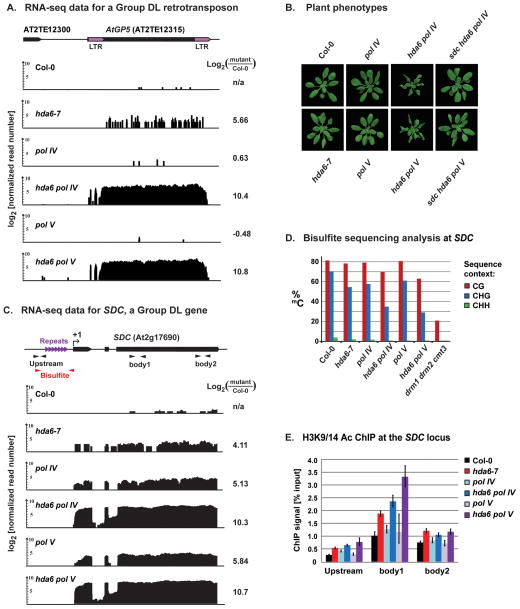
Figure 2. Molecular and phenotypic consequences of “double-locked” silencing involving HDA6 and Pol IV/V.
(A) AtGP5 retrotransposon RNA-seq data. Long terminal repeats (LTRs) flank the transposable element body (see diagram). Vertical black bars denote polyA+ RNA read numbers (log2 units), normalized to total mapped reads, in wild-type Col-0, homozygous hda6-7, pol IV and pol V mutants and the double mutants, hda6 pol IV and hda6 pol V. Numbers at right are RPKM (reads per kilobase per million) log-ratios, comparing expression in each mutant to Col-0.
(B) Phenotypes of 21-day-old plants: wild-type Col-0, or the indicated mutants (see also Figure S2).
(C) Diagram of the SDC locus (At2g17690), showing upstream tandem repeats (purple arrows), the Pol II transcription start site (+1), and PolyA+ RNA-seq data plotted on a log2 scale (black). PCR amplicons for chromatin immunoprecipitation (ChIP) and bisulfite sequencing (black and red arrows, respectively) are indicated.
(D) Bisulfite sequencing analysis of DNA methylation upstream of +1 (red amplicon, panel C). Data from triple mutant drm1 drm2 cmt3 serves as a hypomethylated control (Henderson and Jacobsen, 2008).
(E) ChIP analysis of Histone 3 Lysine 9/14 acetylation at three positions in the SDC locus. Error bars show the propagated standard error of the mean from three replicates.