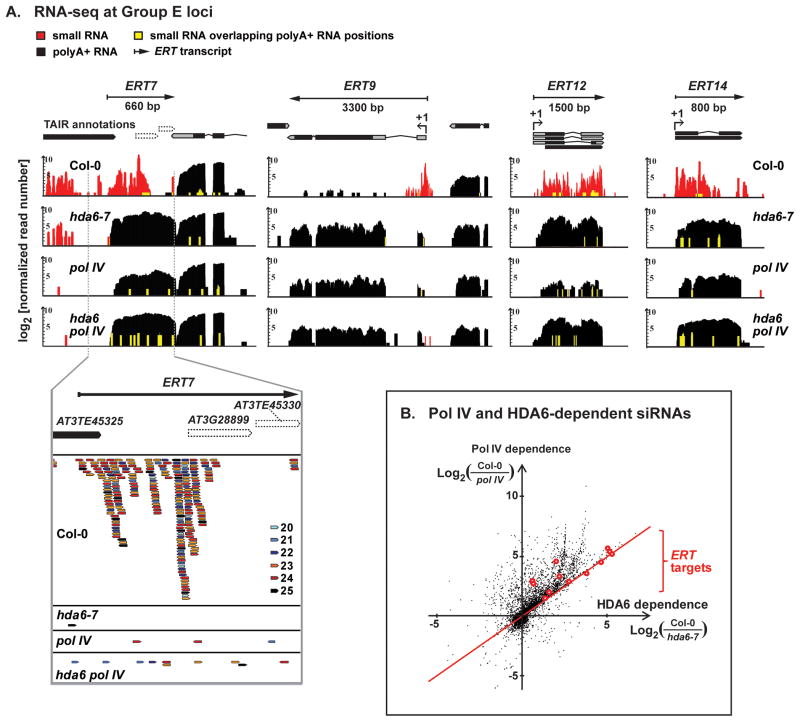
Figure 4. HDA6 facilitates Pol IV-dependent siRNA biogenesis.
(A) RNA-seq profiles at three Group E targets. Data tracks show polyA+ RNA reads (black vertical bars) and small RNA reads (red and yellow vertical bars; yellow denotes positions where siRNA and poly A+ reads overlap) in wild-type (Col-0), hda6-7, nrpd1-3 (pol IV), and the double mutant, hda6 pol IV. Read counts (log2 units) were normalized by total mapped reads. Analogous data for Group DL loci are in Figure S4.
(B) Scatter plot showing the relative dependence of 24 nt siRNA production on Pol IV and HDA6, genome-wide. Each data point represents a TAIR10 gene locus, with the x-axis indicating the log-ratio of small RNA read density in Col-0 relative to hda6-7, and the y-axis indicating the log-ratio of small RNA read density in Col-0 relative to pol IV. Group E loci are marked in red.