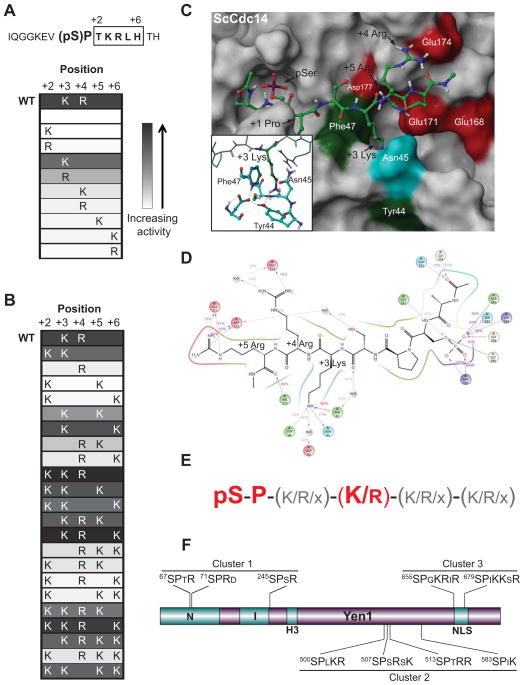
Figure 1. Refinement of Cdc14 enzymatic specificity.
A–B) The peptide sequence from Pds1 was synthesized with a central phosphoserine (pS), and variants contained different combinations of basic amino acids in the +2 to +6 region (boxed). WT, wild-type. Heat map cells show positions and identity of basic residues, Lys (K) or Arg (R), in each peptide. Shading indicates relative activity (white = low, black = high) in Cdc14 assays conducted under steady-state conditions that approximate kcat/Km. C) Representative frame from MD simulation of the ScCdc14 homology model (surface representation) with bound optimal peptide substrate Ac-A(pSer)PSKRR-NHMe (ball and stick). Residues that contact the substrate KRR are colored (red, acidic; green, hydrophobic; cyan, polar neutral) and labeled in white. Key residues of the substrate are labeled in black. Inset highlights the local arrangement of +3 Lys engaged in cation-pi contacts with Tyr44 and Phe47, stabilized by hydrogen bonding (dashed lines). See also Fig. S1. D) 2-dimensional representation of stable (≥ 40% of simulation time) contacts (direct or water-bridged) between peptide (black) and ScCdc14 residues obtained from an MD experiment. Percentages represent the fraction of time that the contact was maintained. Purple arrows are hydrogen bonds, red lines are cation-pi interactions, residue colors are as in panel C with basic residues in blue. E) Summary of Cdc14 substrate features used for substrate prediction. Essential requirements are in large red font. See additional information in Tables S1-S3. F) Diagram of relevant Yen1 protein features. All nine Ser-Pro Cdk site positions with downstream basic residues are shown and divided into the 3 clusters used for the purpose of mutagenesis throughout the paper. N, I, and H3 (helix-hairpin-helix) are conserved domains of the XPG nuclease family.