Figure 1. On-target genome editing activities of truncated gRNAs and Cas9 nuclease in human cells.
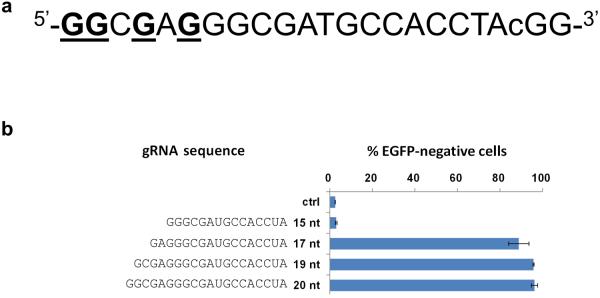
(a) Target site in the EGFP gene used to assess the activities of gRNAs that are truncated at their 5' end. Note that Gs near the 5' end (underlined) enable gRNAs with complementarity lengths of 15, 17, 19 and 20 nt to be expressed from a U6 promoter in human cells. The degenerate base of the protospacer adjacent motif (PAM) sequence (in this case, a C nucleotide) is shown in lowercase.
(b) Efficiencies of EGFP disruption in human cells mediated by Cas9 and gRNAs bearing variable length complementarity regions (shown in nts) for the target site of (a). Ctrl = control gRNA lacking a complementarity region. Error bars indicate standard errors of the mean (s.e.m.), n = 2.
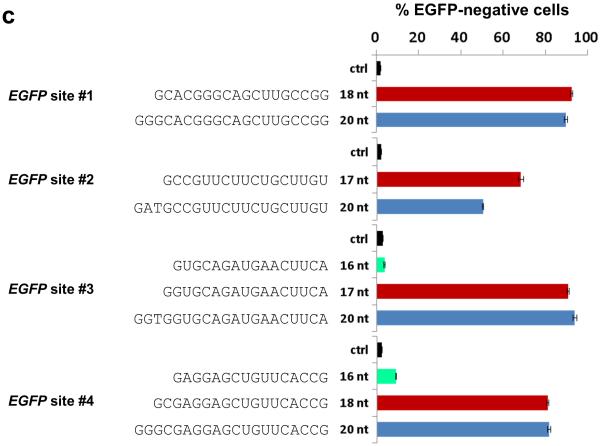
(c) Efficiencies of EGFP disruption in human cells mediated by Cas9 and full-length or shortened gRNAs for four target sites in the EGFP reporter gene (sites #1 – #4). Lengths and sequences of the gRNA complementarity regions are shown. Ctrl, control gRNA lacking a complementarity region; nt, nucleotides. Error bars represent s.e.m., n = 2.
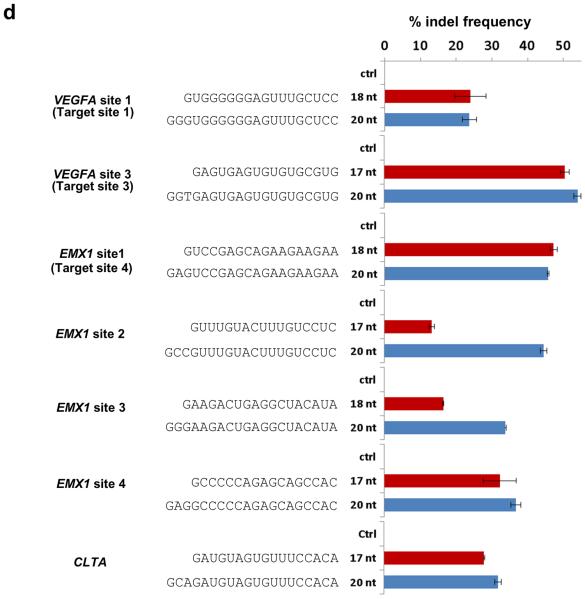
(d) Efficiencies of targeted indel mutations introduced at seven different human endogenous gene targets by matched standard and tru-RGNs. Lengths and sequences of gRNA complementarity regions are shown. Indel frequencies were measured by T7EI assay. Ctrl = control gRNA lacking a complementarity region. Three of the target sites used are named as previously described4: VEGFA site 1 (aka Target site 1), VEGFA Site 3 (aka Target Site 3), and EMX1 Site 1 (aka Target Site 4). VEGFA= vascular endothelial growth factor A, EMX1= empty spiracles homolog 1. Error bars represent s.e.m., n = 2.
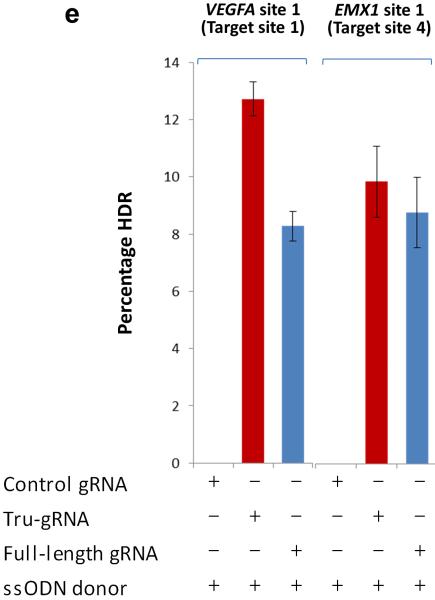
(e) Efficiencies of precise alterations mediated by homology-directed repair (HDR) using single strand oligonucleotides (ssODN) at two endogenous human genes (VEGFA and EMX1) with either full-length RGNs or tru-RGNs. The percentage of alleles in which HDR had occurred (Percentage HDR) was measured using a BamHI restriction digest assay (Online Methods). Control gRNA = control gRNA lacking a complementarity region. Error bars represent s.e.m., n = 2.