Figure 2. tru-gRNAs exhibit enhanced specificities and function efficiently with Cas9 nuclease and paired Cas9 nickases in human cells.
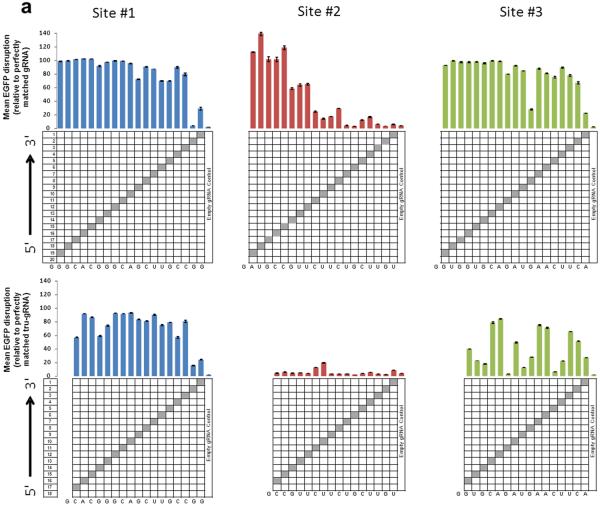
(a) Activities of RGNs targeted to three sites in EGFP using full-length (top) or tru-gRNAs (bottom) with single mismatches at each position (except at the 5'-most base which must remain a G for efficient expression from the U6 promoter). Grey boxes in the grids show the positions of Watson-Crick transversion mismatches. Empty gRNA control is a gRNA lacking a complementarity region. RGN activities were measured using the EGFP disruption assay and values shown represent the percentage of EGFP-negative observed relative to an RGN using a perfectly matched gRNA. Experiments were performed in duplicate and means with error bars representing s.e.m. are shown. Nucleotides present at each position of the gRNA complementarity region are shown at the bottom of each panel.
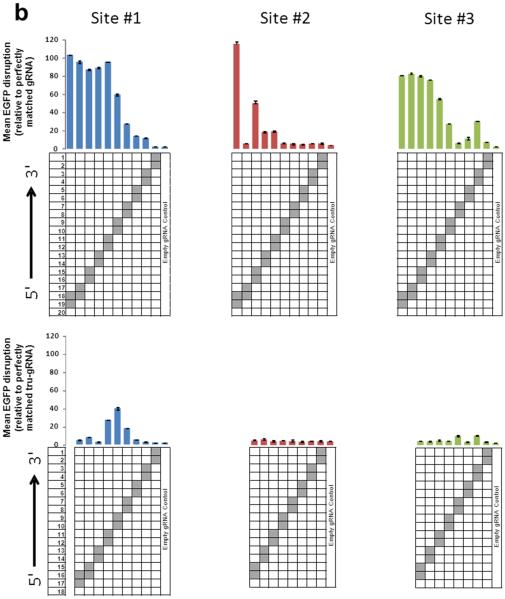
(b) Activities of RGNs targeted to three sites in EGFP using full-length (top) or tru-gRNAs (bottom) with adjacent double mismatches at each position (except at the 5'-most base which must remain a G for efficient expression from the U6 promoter). Data presented as in (a).
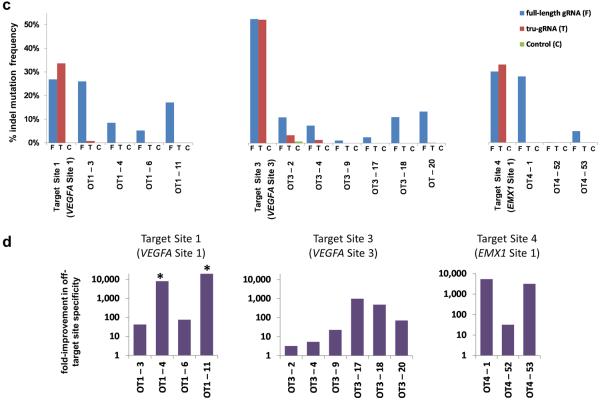
(c) The percentage of sequencing reads containing on- and off-target indel mutations induced by RGNs targeted to three different endogenous human gene sites as measured by deep sequencing. Indel frequencies are shown for the three target sites from cells in which targeted RGNs with a full-length gRNA, a tru-gRNA, or a control gRNA lacking a complementarity region were expressed. Sequences of the on-target and off-target (OT) sites for full-length and tru-gRNAs are shown in Table 1. The scale of the y-axis is the same for all three panels. Absolute counts of indel mutation data used to make these graphs can be found in Supplementary Table 2. On-target sites are named as previously described4 and as in Fig. 1d.
(d) Fold-improvements in off-target site specificities of three tru-RGNs compared to matched standard RGNs. Values shown represent the ratio of on-target to off-target activities for tru-RGNs divided by the ratio of on-target to off-target activities of standard RGNs for the off-target sites shown, calculated using the data from (c) and Supplementary Table 2. For the sites marked with an asterisk (*), no indels were observed with the tru-RGN and therefore the values shown represent conservative statistical estimates for the fold-improvements in specificities for these off-target sites (see Online Methods).
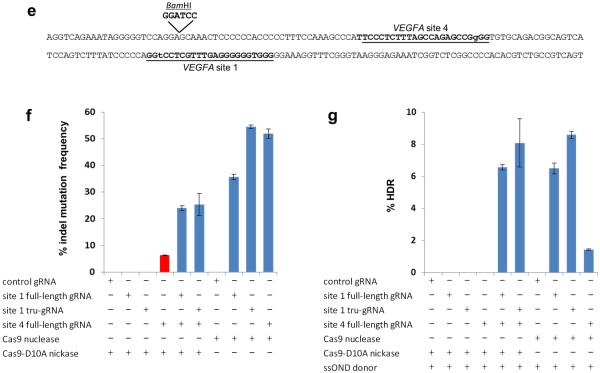
(e) Schematic illustrating locations of VEGFA sites 1 and 4 targeted by gRNAs and variant Cas9 nickases to generate paired double nicks. Target sites for the full-length gRNAs are underlined, with the first base in the PAM sequence shown in lowercase. Location of the BamHI restriction site inserted by homology-directed repair (HDR) with a ssODN donor is shown.
(f) Substitution of a full-length gRNA for VEGFA site 1 with a tru-gRNA does not reduce the efficiency of indel mutations observed with a paired full-length gRNA for VEGFA site 4 and Cas9-D10A nickases. Control gRNA used is one lacking a complementarity region. The frequency of indel mutations induced by the VEGFA site 4 gRNA alone with Cas9 nickase is highlighted in red.
(g) Substitution of a full-length gRNA for VEGFA site 1 with a tru-gRNA does not reduce the efficiency of BamHI sequence alterations introduced with a paired full-length gRNA for VEGFA site 4, Cas9-D10A nickase, and an ssODN donor template. Control gRNA used is one lacking a complementarity region.