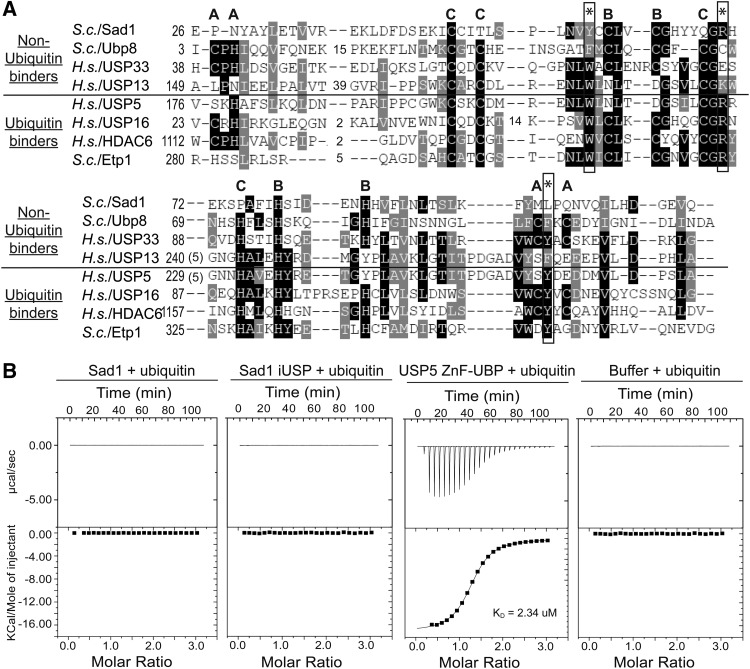
FIGURE 2.
Sad1 does not bind ubiquitin in vitro. (A) A primary sequence alignment of ubiquitin binding and nonubiquitin binding Homo sapiens and S. cerevisiae ZnF-UBP domains. The WRY motif residues that are essential for ubiquitin binding in USP5 are annotated with asterisks; in Sad1, they appear as YRL. The residues that comprise the three zinc fingers found in ZnF-UBPs are denoted A, B, and C; Sad1 only possesses the B site zinc finger. Conservation is indicated by shades of gray, and gaps in the alignment are represented by dashes. (B) We titrated 1 mM of ubiquitin into an ITC receptor cell containing either 50 µM full-length Sad1, Sad1-iUSP, USP5 ZnF-UBP, or buffer at 25°C. The data from the USP5 ZnF-UBP titration with ubiquitin were fit to a single exponential in good agreement with one binding site and a dissociation constant of 2.3 µM. ITC curve-fitting and data analysis were performed with the MicroCal-enabled Origin software.