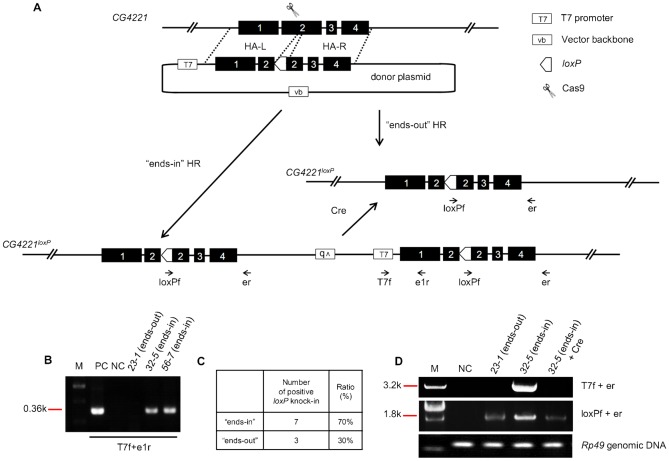
Fig. 5. Removal of “ends-in” recombination resultant from CG4221 mutagenesis.
(A) “Ends-in” and “ends-out” homologous recombination (HR) were generated via the HDR pathway in the process of CG4221 mutagenesis, leading to either two copies of the CG4221 mutations or one copy of the CG4221 mutation in the fly genome, respectively (see also supplementary material Fig. S11). The Cre recombinase was introduced into the “ends-in” line to remove the duplicated copy of CG4221 by flipping out the DNAs between the two loxP sites, converting the “ends-in” into the “ends-out” events. The short arrows with names indicate the primers used in panels B,D. (B) PCR assays to distinguish the “ends-in” and “ends-out” HR events occurred during CG4221 mutagenesis. The genomic DNAs of heterozygous F1 lines 23-1, 32-5 and 56-7 were used for PCR assays. T7f and e1r detect a corresponding 0.36 kb “ends-in” band, while nothing for the “ends-out”. (C) “Ends-in” and “ends-out” HR ratio of CG4221 mutagenesis, based on the PCR analyses as described in panel B. (D) Molecular confirmation of the removal of the “ends-in” products. Line 32-5 was selected as an example to show successful removal of additional sequences between the two loxP sites by Cre recombinase, as indicated by the loss of the band amplified with T7f and er. Primer pairs used for PCRs are indicated on the right of the gel. Rp49 genomic primers were used for genomic DNA quality control. See the legends to Fig. 1 for the common elements or labels that are not explained here.