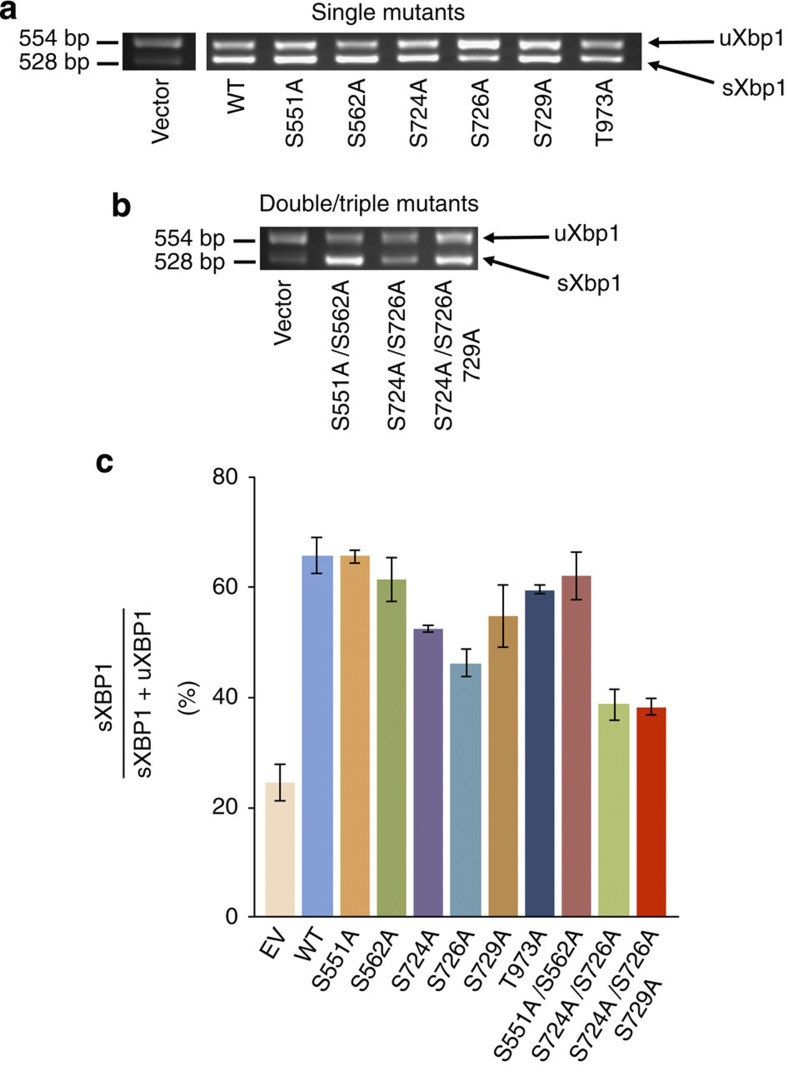
Figure 4. Mutation of phosphorylation sites inhibits splicing in cells.
(a) Xbp1 splicing in HT 1080 human cell line. Individual phosphorylation mutants were co-transfected into HT1080 cells and subjected to tunicamycin for 2 h to induce ER stress. RT–PCR of sXbp1 and uXbp1 was run on 3% agarose gel and visualized by gel red DNA stain. Empty vector was used as control, and all experiments were repeated in triplicate. (b) Xbp1 splicing using sets of mutations mimicking groups of phosphorylations within Ire1. S551A/S562A represents L-phos mutations, S724A/S726A replicates in vitro re-phosphorylation of dephos Ire1 with ATP, S724A/S726A/S729A triple mutant represents all activation loop phosphorylations. (c) Quantification of inhibition of Xbp1 splicing in vivo of each individual phosphorylation mutant and sets of mutants in HT1080 cells, with empty vector (EV) as control. Splicing is clearly reduced in all activation loop mutants and especially in the double and triple mutants recapitulating the in vitro findings. Linker region or L-phos single and double mutants seem not to have an impact on splicing, whereas T973A seems to have a small impact. Error bars represent mean±s.e.m., n=3.