Figure 1.
The Unwinding Rate of T7 Helicase Depends on Base Pair Stability
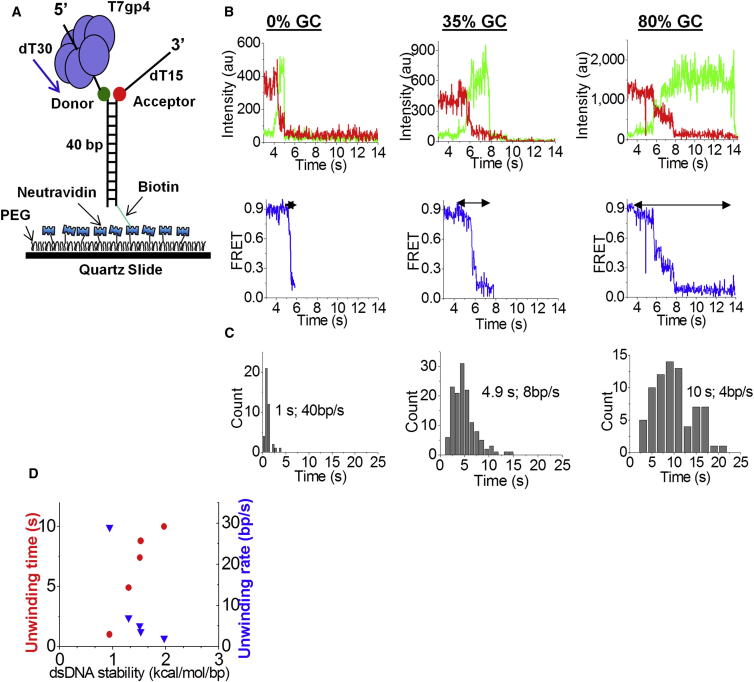
(A) T7 gp4 was loaded on a 40 bp DNA with (dT)n tails containing donor (Cy3) and acceptor (Cy5) dyes, and bound to a PEG-coated surface via biotin-neutravidin interaction.
(B) Cy3 and Cy5 intensity traces during unwinding for one molecule on mixed, 100% AT, and 80% GC sequences (top panel); calculated FRET efficiency versus time for the fluorescence intensity traces (bottom panel).
(C) Dwell-time histograms during unwinding. The arrows on the FRET traces indicate the intervals at which the dwell times were measured; 50 molecules were used to build the histograms. The data are representative of multiple experiments.
(D) Unwinding time (red circles) and rate (blue triangles) versus base pair stability. Five sequences were used to plot the graph (Table S1).
See also Figure S1.