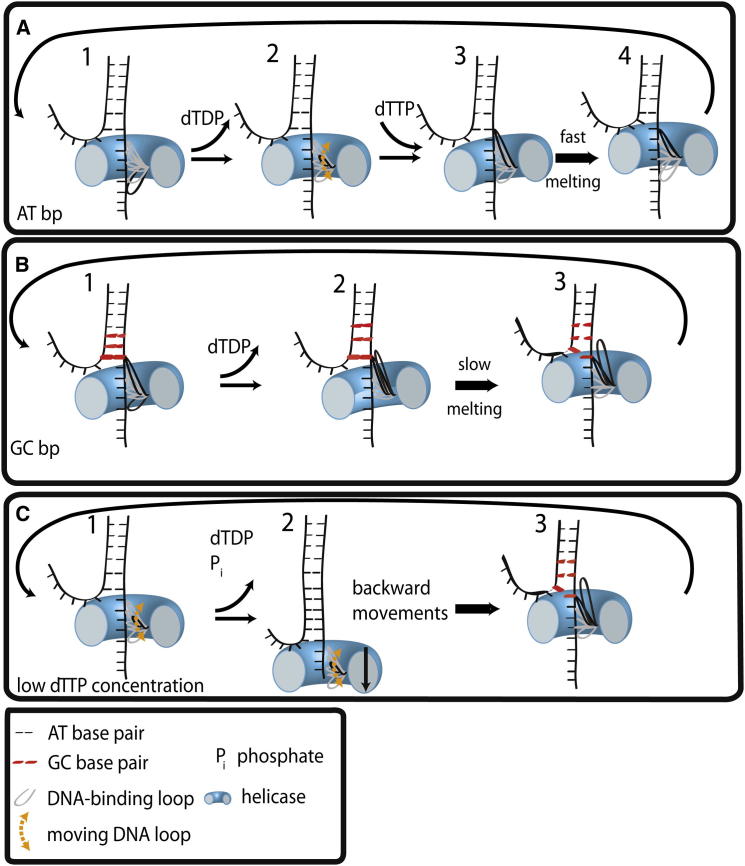
Figure 5.
T7 Helicase Unwinding Model
(A) dsDNA unwinding of AT base pairs. Only the DNA-binding loop of the rearmost position is shown, which is released upon dTDP release. Next, dTTP binds and the released loop binds the DNA at the foremost position. The fast melting of AT base pairs enables the enzyme to move forward.
(B) dsDNA unwinding of GC base pairs. Slow melting of GC base pairs slows down the enzyme. When the enzyme encounters GC base pairs, reengagement of the disengaged loop with the DNA ahead is hindered due to steric mismatch between dsDNA and central helicase pore. Subsequent dTTP hydrolysis and product release by the next rearmost subunits build enough strain in the system to eventually cause 2–3 bp unwinding in a burst.
(C) dsDNA unwinding at low dTTP concentrations. The DNA-binding loop does not readily bind to the DNA because the subunit remains in the dTTP unbound state. Subsequent dTTP hydrolysis and dTDP release at other subunits leads to the detachment of all of the DNA-binding loops and backward movements by the enzyme, and the DNA rezips. Once a new set of contacts is created by the enzyme on the tracking strand, the enzyme can move forward and unwind DNA.