Figure 5.
Structural, Biophysical, and Biochemical Characterization of Oncogenic RET M918T
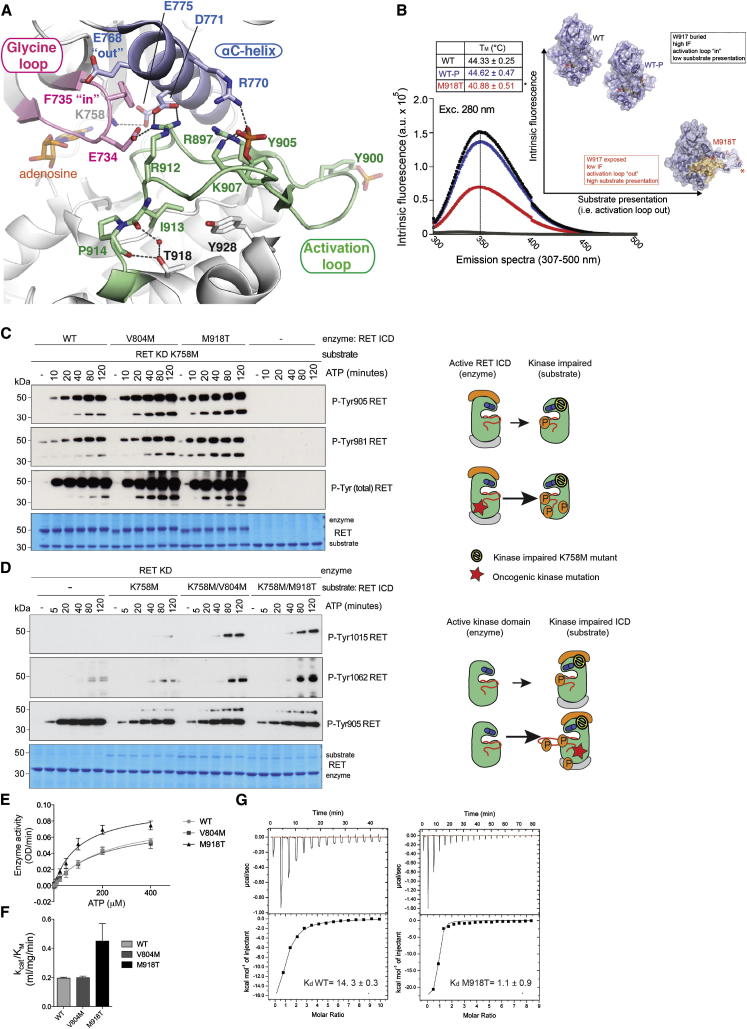
(A) Crystal structure of oncogenic RET M918T KD at 2.1Å resolution (PDB ID code 4CKI). Closed-up view shows the GRL in the closed conformation. Main secondary structure components, ligand adenosine, and important residues are depicted with discrete colors.
(B) Biophysical characterization of RET ICD WT and oncogenic M918T. IF (arbitrary units [a.u.]) emission spectra (307–500 nm, excitation at 280 nm) of RET ICD WT, phosphorylated RET ICD WT, and RET M918T. Left inset: shows the melting temperatures for the indicated proteins, showing a significantly lowered melting temperature for oncogenic RET M918T. Right inset: depicts our proposed model of enhanced substrate presentation in trans by an “AL-out” conformer. Asterisk (∗) denotes W917.
(C) WB analysis of autoP rescue experiments in trans of catalytically impaired RET K758M KD (substrate, 2 μM) by active RET ICD (2.5 μM) WT, V804M, and M918T using the indicated antibodies. Total levels of RET (both enzyme and RET K758M substrate) were monitored by Coomassie staining. Time course with ATP (5 mM) and MgCl2 (10 mM) for 0–120 min, n = 2.
(D) WB analysis of autoP rescue experiments in trans of catalytically impaired RET K758M, RET K758M/V804M, or K758M/M918T ICD (1 μM) by RET KD (2.5 μM) using the indicated antibodies. Total levels of RET were monitored by Coomassie staining. Time course with ATP (5 mM) and MgCl2 (10 mM) for 0–120 min, n = 3.
(E) Enzymatic assay performed with purified recombinant RET ICD WT, V804M, or M918T (2.5 μM) incubated with increasing concentrations of ATP (0–400 μM) using 8 mg/ml ABL peptide (EAYAAPFAKKK). Data represent the mean (OD/min) ± SE, n = 4.
(F) Catalytic efficiency constants (kcat/KM) of (E).
(G) Isothermal titration calorimetry profiles of ATP for RET ICD WT and M918T at pH 7.65 at 20°C. The plot of the heat released (kcal) per mol of ATP added, against the molar radio of ATP to protein is depicted. Note that a minor proportion of ATP may be converted to ADP via autoP. The data were fitted to a single sequential-binding site(s) model, and the solid lines represent the best fit.
See also Figures S4, S5, and Table S1.