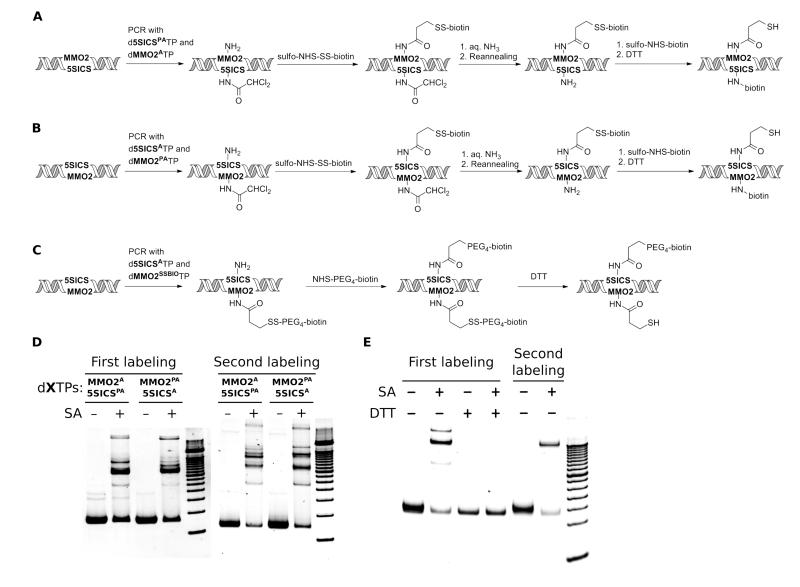
Figure 5.
Post-amplification DNA labeling with two different functional groups, using (A) d5SICSPA-dMMO2A, (B) d5SICSA-dMMO2PA, and (C) d5SICSA-dMMO2SSBIO. (D) Efficiencies of first and second labeling of DNA from containing d5SICSPA-dMMO2A and d5SICSA-dMMO2PA. First labeling efficiencies are 61% for d5SICSPA-dMMO2A and 66% for d5SICSA-dMMO2PA, and second labeling efficiencies are 85% for d5SICSPA-dMMO2A and 78% for d5SICSA-dMMO2PA. A 100 bp ladder is loaded in the rightmost lane of each gel. (E) First and second labeling efficiencies of DNA containing d5SICSA-dMMO2SSBIO. The first labeling efficiency is 85% in the absence of DTT and 0% in the presence of DTT (due to linker cleavage). The second labeling efficiency is 68%. A 50 bp DNA ladder is loaded in the rightmost lane. In (D) and (E), the faster migrating, strong band corresponds to dsDNA, while the slower migrating band corresponds to the 1:1 complex between dsDNA and streptavidin. The faint and more slowly migrating bands correspond to higher order complexes of dsDNA and streptavidin.