Figure 3.
Modulation of Origin Efficiencies by Altering Levels of the Replication Machinery Induces Specific Changes in Rad51 Binding
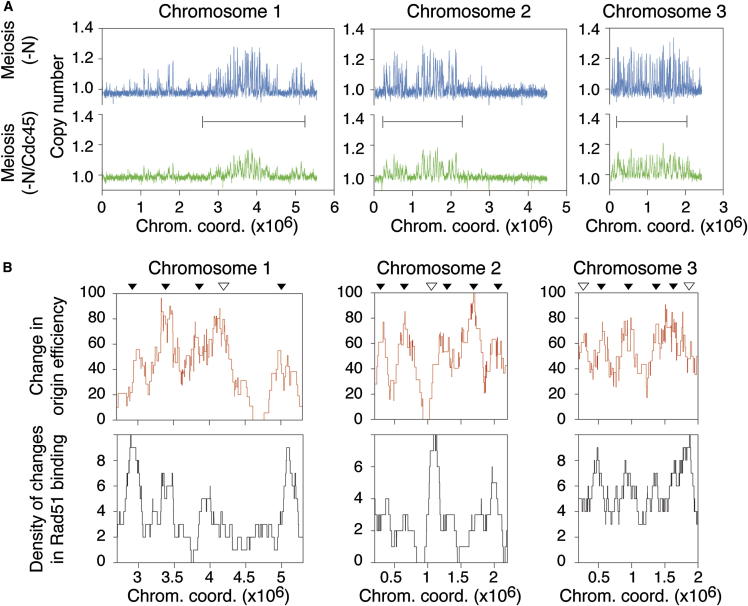
(A) Decrease in origin efficiencies across the genome as a result of the dominant-negative effect of cdc45 overexpression in nitrogen starvation conditions. Replication profiles in strains induced to undergo meiosis after nitrogen depletion (−N) or after nitrogen depletion with altered Cdc45 levels (−N/Cdc45; see Experimental Procedures). The lines in each of the three graphs delineate the regions analyzed in Figure 3B.
(B) Genome-wide analysis of changes in origin efficiencies and Rad51 binding in −N versus −N/Cdc45 conditions. As there are large areas of the genome with few or no apparent active origins above our detection threshold in both the −N and −N/Cdc45 conditions, only regions where there are sufficient origin densities and efficiencies for reliable analysis were considered (Figure S4A). Top panels: changes in origin efficiencies over continuous ∼200 kb windows (analysis as performed for Figure 2A). The difference between the two conditions is shown ([−N] − [−N/Cdc45]). Bottom panels: density of increases in Rad51 binding along the genome in −N compared to −N/Cdc45 conditions (analysis as in Figure 2A). The locations of (1) sites of Rad51 binding present only in −N conditions and (2) sites for which (−N) / (−N/Cdc45) ≥ 2 for the peak values were identified and combined over ∼200 kb windows. Consistent with the lower origin efficiencies across the genome in −N/Cdc45, we found very few Rad51 binding sites that are unique to or increased in −N/Cdc45 compared to −N (Figure S4B). In both the top and bottom panels, the abscissa represents the moving average of the chromosome coordinates for the corresponding ∼200 kb windows. Black triangles indicate a correspondence of the major peaks between the origin efficiency and Rad51 binding results, and white triangles denote differences.