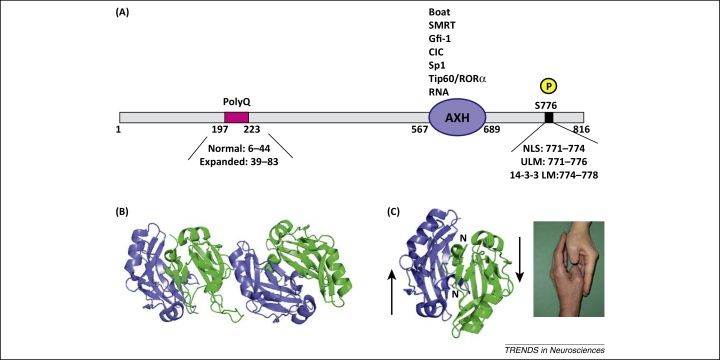
Figure 2.
The structure of Atx1. (A) A schematic representation of the Atx1 architecture. The protein is represented as a horizontal bar on which the position of the polyQ motif, the AXH domain and S776 are indicated. The interactions formed with various cellular partners are also listed near the interacting motif. (B) The crystal structure of the AXH domain (PDB accession code 1oa8, [21]). In the crystallographic asymmetric unit, there are two dimers, which form a dimer of dimers (indicated in the figure with alternated colors). (C) Structure of the dimer rotated by 45 degrees around the axis perpendicular to the plane as compared to the view in (B). The overall arrangement is antiparallel with the termini being sandwiched in the dimer interface like in two touching left hands. The symmetry is however incomplete and the individual protomers differ by local details. Most of the differences are grouped at the N termini. Abbreviations: Atx1, ataxin-1; CIC, Capicua; NLS, nuclear localization signal; PDB, Protein Data Bank; polyQ, polyglutamine; UHM, U2AF (U2 auxiliary factor) homology motif; ULM, UHM ligand motif.