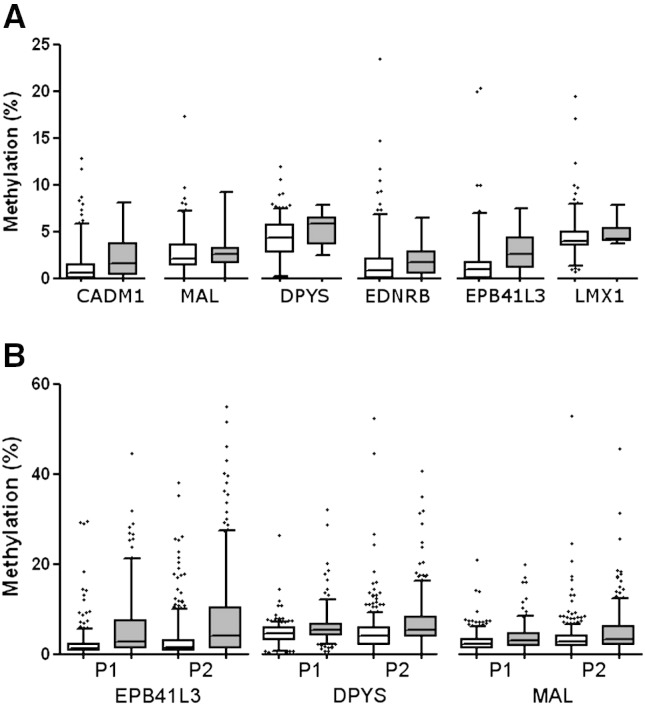
Fig. 1.
The distribution of methylation in HR-HPV negative and positive samples. A) Comparison of methylation distribution of 6 genes in 207 HR-HPV negative controls (white) and 10 CIN2/3 (grey) in the P1 study. EPB41L3 (p = 0.023) and DPYS (p = 0.043) showed elevated methylation in CIN2/3 while CADM1 (p = 0.051), MAL (p = 0.810) and LMX1 (p = 0.177) did not. B) Comparison of EPB41L3, DPYS and MAL methylation distribution in HR-HPV positive <CIN1 controls (white) and CIN2/3 (grey) from P1 and P2 cohort. Significantly elevated methylation was observed in CIN2/3 group for all three genes in both studies (p < 0.0001). Numbers of patients in each group are available in Supplemental table S1. For better visualisation of the low methylation results, 2 outliers are not shown, one at 70% EPB41L3 methylation in a normal tissue from P2 and one at 91% DPYS methylation in a normal tissue from P1. Whiskers of the boxplot mark the 5th and 95th percentiles, the box 25th percentile, median and 75 percentile, while extreme values are shown by (•).