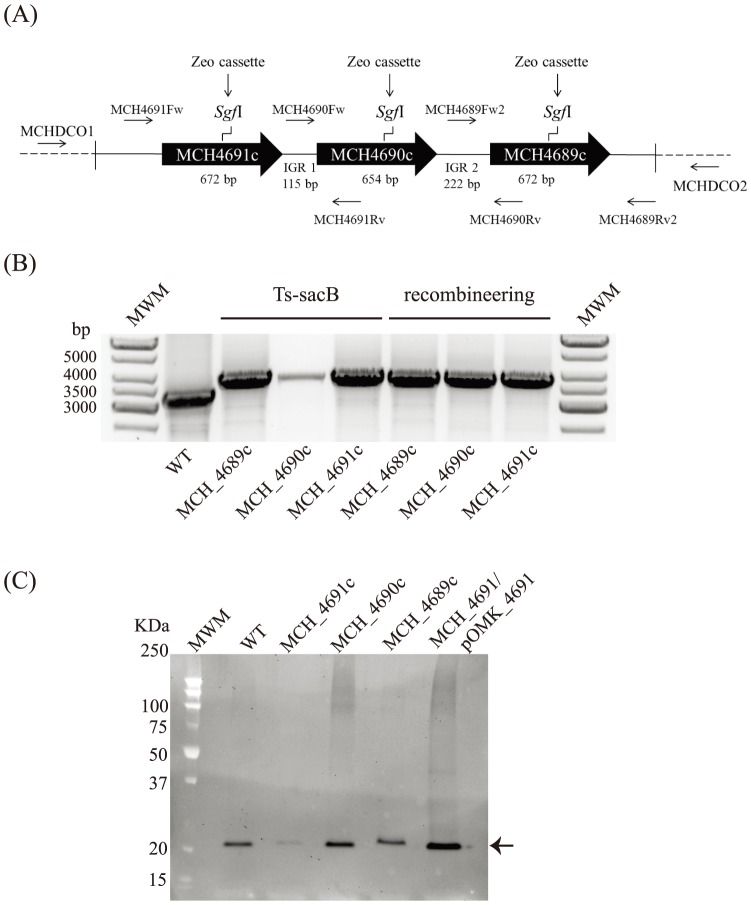
Figure 1. Gene replacement at the MCH_4689c, MCH_4690c and MCH_4691c porin loci of M. chelonae ATCC 35752 using the Ts-sacB and recombineering systems.
(A) Porin gene cluster of M. chelonae ATCC 35752. The positions of the primers used to generate the allelic exchange substrates and analyze the candidate mutants are indicated. IGR1 and IGR2 represent the intergenic regions. (B) Candidate mutants obtained for each of the porin genes using the Ts-sacB or the recombineering systems were analyzed by PCR as described under Materials and Methods and confirmed by sequencing the regions flanking the resistance cassette. The expected size of the PCR fragments is 3.3 kb for the wild-type parent strain and 3.8 kb for the knock-out mutants. MWM, molecular weight marker. WT, wild-type. (C) Immunoblot analysis of porin production in the wild-type, mutant and complemented mutant strains. Strains were grown in 7H9-OADC-Tween 80 broth at 30°C to mid-log phase (OD600 = 1) and porins were selectively extracted from whole cells at 100°C using 0.5% n-octylpolyoxyethylene as a detergent as described [44]. Protein samples prepared from the same amount of cells for each strain were denatured by boiling in 80% DMSO followed by acetone precipitation [23]. Denatured proteins were loaded volume to volume, separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and porins were detected using rabbit antiserum to purified MspA [23]. Immune complexes were detected by chemiluminescence (Pierce, ELC) and semi-quantified using the Image Lab software (Biorad).