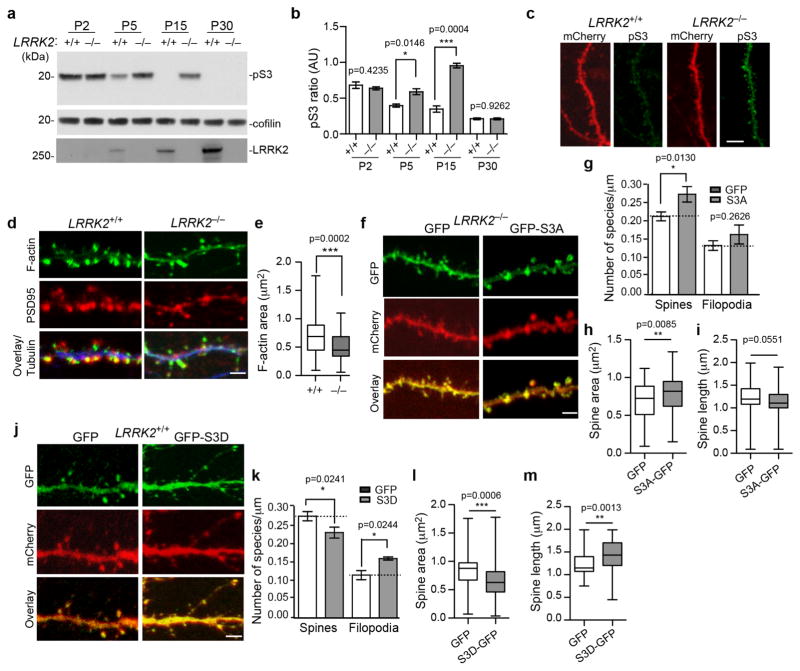
Figure 2. Abnormal phosphorylation of cofilin in LRRK2−/− neurons.
(a) Western blot analysis of forebrain homogenates from LRRK2+/+ and LRRK2−/− mice. The full-length images are in shown in Supplementary Fig. 2e. (b) The pS3 cofilin levels normalized against total cofilin levels (n=3 per genotype). Data represent mean±SEM, Unpaired t-test, t(4)=0.8906, 0.1047, 11.11, and 4.117, respectively. *p<0.05, ***p<0.001. (c) Representative images of spines and their parental dendrites of cultured LRRK2+/+ and LRRK2−/− hippocampal neurons transfected with mCherry (red) and stained with pS3 cofilin (green). Scale bar: 10μm. (d) Staining of F-actin (green), PSD95 (red), and βIII-tubulin (blue) in dendritic spines and corresponding dendrites of LRRK2+/+ and LRRK2−/− hippocampal neurons at 15DIV. Scale bar: 5μm. (e) F-actin area staining in the dendritic spine of LRRK2+/+ and LRRK2−/− hippocampal neurons (n=6–8 neurons per genotype; n=85–90 spines per genotype). Data represent mean ± SEM. Unpaired t-test, ***p<0.001. (f, j) Spines and dendritic regions of LRRK2−/− (f) and LRRK2+/+ (j) hippocampal neurons transfected with GFP, GFP-S3A, and GFP-S3D cofilin plasmids, along with mCherry at 15DIV. Scale bar: 5μm. (g, k) Density of dendritic spines and filopodia was quantified from LRRK2−/− and LRRK2+/+ neurons. (h–i, l–m) Bar graphs showing average head spine diameter and dendritic spine length of transfected LRRK2−/− (h, i) and LRRK2+/+ neurons (l, m) (LRRK2−/−, nGFP=73 spines, nS3A-GFP=80 spines; LRRK2+/+ nGFP=101 spines, nS3D-GFP=79 spines; Four to seven neurons per genotype). Data represent mean ± SEM. Unpaired t-test, *p<0.05, **p<0.01, ***p<0.001