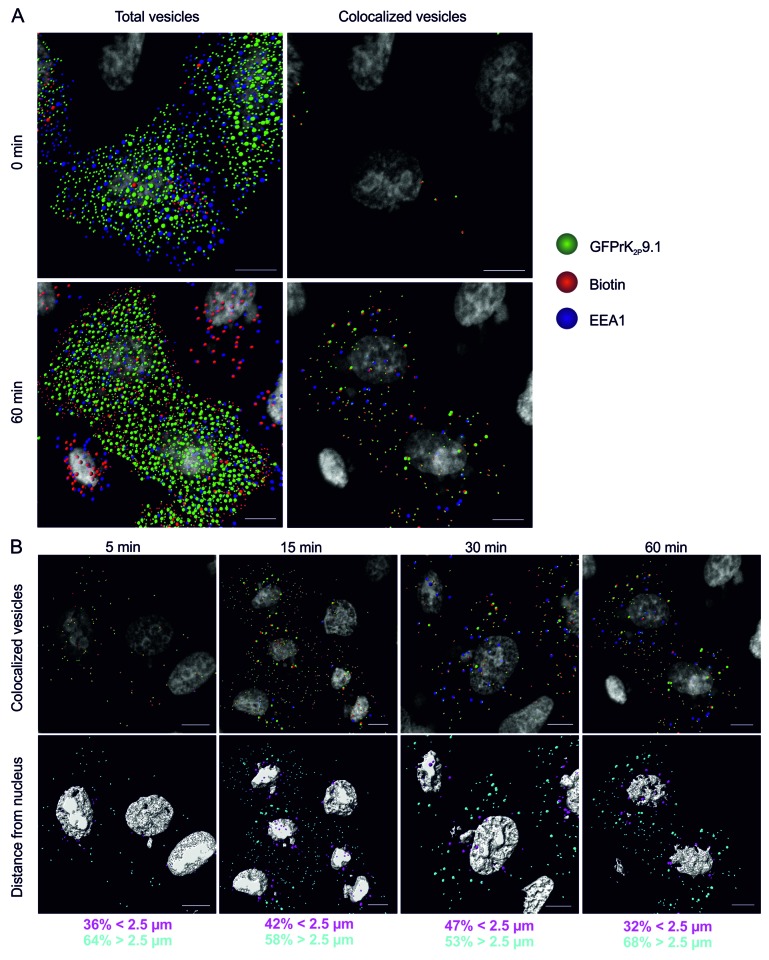
Figure 2. Endocytosis of GFPrK2P9.1. (A) HeLa cells transiently expressing GFPrK2P9.1 (green) were surface-biotinylated on ice and either immediately stripped to remove the external biotin and fixed (upper images, 0 min) or warmed to 37 °C for 60 min (lower images, 60 min) before stripping and fixing. Cells were stained with streptavidin-Alexa Fluor 546 (red) to reveal internalized biotin and with anti-EEA1 (blue). Confocal z-stacks were analyzed by Imaris to detect 0.6 μm and 1.2 μm vesicles, as in Figure 1. Total vesicles: all the spots detected by Imaris software. Colocalized vesicles: all the colocalized GFPrK2P9.1-biotin spots (green and red) and triple colocalized GFPrK2P9.1, biotin and EEA1 spots (green, red and blue). Bar represents 10 μm. (B) HeLa cells from the time-course experiment shown in (A) were analyzed after 5-, 15-, 30- and 60-min incubation at 37 °C to identify internalized GFPrK2P9.1. Upper images show colocalized GFPrK2P9.1-biotin spots (green and red) and triple colocalized GFPrK2P9.1, biotin and EEA1 (green, red and blue). Lower images show the same cells, but with the vesicles color-coded according to distance to the nucleus. Magenta vesicles are nearer than 2.5 μm and cyan vesicles are further than 2.5 μm from the nearest nucleus. Numbers represent the proportion of vesicles in each distance category. Scale bars: 10 μm.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.