Figure 7.
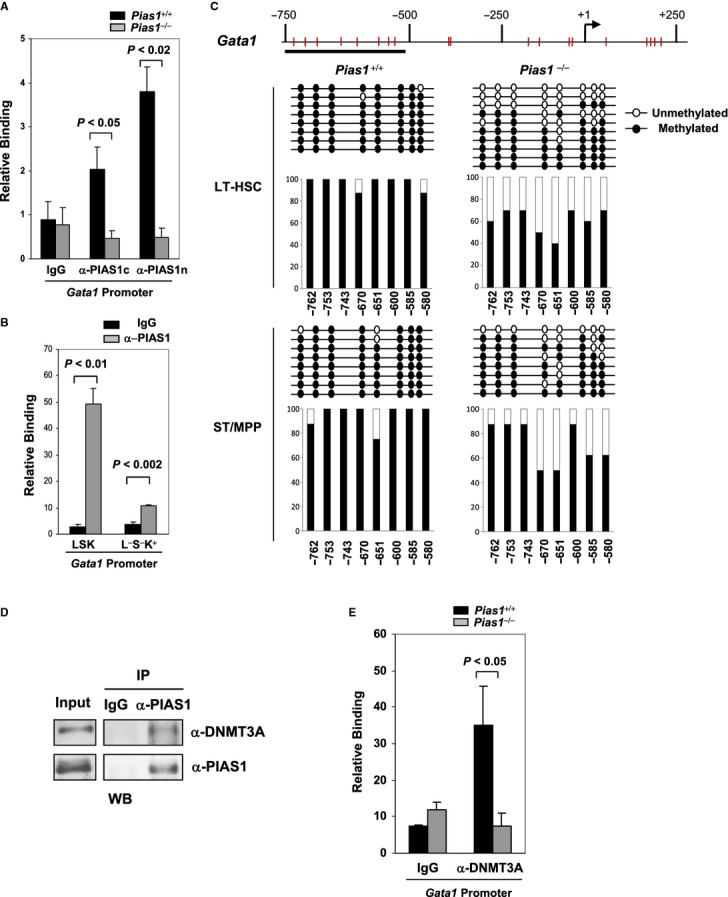
- Chromatin immunoprecipitation (ChIP) assays were performed with cell extracts from WT or Pias1−/− BM, using 2 independent antibodies against either the C-terminal (α-PIAS1c) or the N-terminal (α-PIAS1n) PIAS1, or IgG as a negative control. Bound DNA was quantified by Q-PCR with specific primers against Gata1 promoter, and normalized with the input DNA.
- Same as in (A) except that FACS-sorted LSK or Lin−Sca1−c-Kit+ (L−S−K+) cells from WT BM were used.
- Methylation analysis of the Gata1 promoter was performed by bisulfite conversion of genomic DNA from FACS sorted long-term hematopoietic stem cells (LT-HSC) and short-term multi-potent progenitors (ST/MPP) as defined in Materials and Methods from WT and Pias1−/− male mice (n = 5). The x axis represents the positions of the CpG sites relative to the transcription start site (+1); the y axis represents the percentage.
- PIAS1 interacts with DNMT3A in BM cells. Co-immunoprecipitation (Co-IP) assays were performed with cell extracts from WT BM, using anti-PIAS1 or IgG, followed by immunoblotting with anti-DNMT3A or a monoclonal anti-PIAS1.
- PIAS1 is required for the recruitment of DNMT3A to the Gata1 promoter. Same as in (A) except that anti-DNMT3A was used for ChIP assays.
Data information: Shown in each panel is a representative of 3 independent experiments (n = 4–6 for each experiment). Error bars represent SEM. P-values were determined by non-paired t-test.
