Abstract
Tissue homeostasis depends largely on the ability to replenish impaired or aged cells. Thus, tissue-resident stem cells need to provide functional progeny throughout the lifetime of an organism. Significant work in the past years has characterized how stem cells integrate signals from their environment to shape regulatory transcriptional networks and chromatin-regulating factors that control stem cell differentiation or maintenance. There is increasing interest in how post-translational modifications, and specifically ubiquitylation, control these crucial decisions. Ubiquitylation modulates the stability and function of important factors that regulate key processes in stem cell behavior. In this review, we analyze the role of ubiquitylation in embryonic stem cells and different adult multipotent stem cell systems and discuss the underlying mechanisms that control the balance between quiescence, self-renewal, and differentiation. We also discuss deregulated processes of ubiquitin-mediated protein degradation that lead to the development of tumor-initiating cells.
Keywords: differentiation, malignancy, proteasome, stem cells, ubiquitin
Stem cells: concepts and definitions
Embryonic stem cells and adult tissue-resident stem cells are of great interest in biology and medicine due to their unique characteristics 1. They have the ability to self-renew, which is defined as the capacity to proliferate while being able to differentiate to downstream cellular types upon proper stimuli from their environment. This unparalleled cellular plasticity of stem cells identifies them as key determinants of tissue equilibrium.
Although progenitor cells also have the ability to self-renew, this is usually a short-term characteristic 2. The long-term self-renewal capacity of stem cells is essential to supply tissues with differentiated progeny throughout the life of the organism. Adult stem cells reside in specialized microenvironments called niches and manifest different degrees of quiescence, depending on the specific organ characteristics. For example, hematopoietic stem cells (HSCs) are dormant 3, whereas mammary stem cells (MaSCs) appear to be cycling 4 and intestinal stem cells (ISCs) proliferate rapidly 5.
Stem cells can divide symmetrically or asymmetrically [6; Sidebar A]. Symmetric cell divisions ensure that all elements are distributed equally between the two identical daughter stem cells, and differentiation—usually of only one of the daughter cells—occurs at a later stage. Asymmetric cell divisions, on the other hand, lead to the unequal division of stem cell components in a process that involves proper positioning of the mitotic spindle 7. As a result, one cell remains a stem cell, whereas the other adopts a different cell fate. Asymmetric divisions also physically displace one daughter cell from its relative position to the niche, leading to its differentiation.
Sidebar A: Regulation of symmetric and asymmetric stem cell divisions by ubiquitylation.
The Drosophila and C. elegans embryos constitute a powerful tool to study the mechanisms of asymmetric cell division during early development. Several ubiquitin-mediated pathways have been recently implicated in these processes. The E3 ligase Neuralized (Neur) has been shown to regulate epithelial cell polarity 211. Neur ubiquitylates the Notch ligand Delta, promoting its internalization. In addition, bearded can inhibit Neur, restricting its activity to the mesoderm and contributing to the establishment of cell polarity. In an analogous function, NEUR also promotes NOTCH DL internalization in the apical zone of the polarized human kidney cell line MDCK 212. However, the specific roles of Neur during mammalian development in vivo and whether this E3 ligase is important in the adult epithelial cells have not been explored yet.
The asymmetric inheritance of cellular components in C. elegans is controlled by the interplay between PIE-1 and MEX-5. PIE-1 represses transcription by promoting the expression of germline-associated genes 213. MEX-5 on the other hand, through activation by ZIF-1 and phosphorylation by PAR-1 214, forms an E3 ligase complex that degrades PIE-1, establishing segregation and anterior–posterior cytoplasm specification 6.
In addition, the E3 ligase SCFSlimb (SCF-βTrcp in mammals) was shown recently to regulate asymmetric division in Drosophila neuroblasts 215. Slimb is able to associate with kinases Sak and Akt, promoting their ubiquitylation and inhibiting ectopic neuroblast formation. Supporting this notion, β-Trcp is often deleted in human gliomas with a simultaneous activation of Akt signaling 216. SCFSlimb was also implicated in the degradation of Oskar in the Drosophila oocyte 217. In the latter case, Par-1 was shown to be the priming kinase, which allows Gsk3 to phosphorylate an Oskar degron in order to allow degradation by SCFSlimb and establish polarity.
These examples demonstrate the importance of ubiquitin-regulating mechanisms in the balance between symmetric or asymmetric stem cell divisions that establish early tissue specification.
Signals from the niche microenvironment are critical in regulating intrinsic stem cell transcriptional programs. Various signaling pathways such as Wnt, Hedgehog, Notch, TGF-β/BMP, and JAK/STAT act in concert to shape the regulatory networks that control cell cycle progression or exit, differentiation, and homeostasis. Disturbing the balance between these signaling pathways can deregulate these processes and lead to tumor formation 8. Thus, the precise control of these pathways, both in stem and in niche cells, is crucial to execute proper developmental programs. The control of protein stability and/or activity by ubiquitylation is essential in the control of the above-mentioned signaling pathways, and its manipulation can either support or alter stem cell properties.
The nuts and bolts of ubiquitylation
The regulation of protein stability is a crucial function in the control of cell plasticity. The ubiquitin-proteasome system (UPS) is a fundamental mechanism to regulate protein stability, quality control, and abundance. Ubiquitylation is a post-translational modification process that results in the covalent conjugation of the small, highly conserved, 76-amino acid protein ubiquitin to lysine residues of substrate proteins through a cascade of enzymatic reactions 9. These events involve the activation of ubiquitin using ATP by E1-activating enzymes, followed by its transfer to E2-conjugating enzymes and finally the formation of an isopeptide bond between ubiquitin and the substrate protein catalyzed by E3 ligases, which confer substrate specificity 10. This cascade can be repeated multiple times resulting in polyubiquitylated substrates, where each ubiquitin moiety is conjugated to the previous one.
Ubiquitin contains seven lysines (K6, K11, K27, K29, K33, K48, and K63), all of which can be acceptors for the next ubiquitin, as can the amino-terminal methionine. As a result, polyubiquitylation can generate substrates tagged with different types of ubiquitin chain, as well as branches of mixed-chain composition 11. These different chain linkages result in different degrees of polyubiquitylated chain compaction, which can mediate diverse cellular outcomes. For example K11-linked chains, which have some degree of structural flexibility, have been implicated in mitotic degradation 12, whereas K63 chains, which have open, linear-like conformations, have been associated with the activation of kinases 13, 14. A well-studied type is the highly compact K48-linked ubiquitin chain, which serves as the canonical signal for degradation by the proteasome 15. Monoubiquitylation and polyubiquitylations have been implicated in regulating virtually all cellular signaling pathways and processes 16, in addition to maintaining proteostasis. The different ubiquitin chains are recognized by ubiquitin-binding domains of “reader” proteins, thereby deciphering this three-dimensional code 17, 18.
The proteasome is a multimeric enzymatic complex that consists of the 20S catalytic core and one of three regulatory particles, 19S, 11S/PA28, or Blm10/PA200. The core contains the catalytic sites for degradation, and the regulatory particle is responsible for substrate recognition, removal of the polyubiquitin chains, unfolding and translocation into the catalytic cavity 19. Regulatory particles contain ubiquitin-binding receptors 20, deubiquitylating enzymes (DUBs) 21, and ATPases 22 in order to perform these functions. The catalytic core has trypsin-, chymotrypsin-, and caspase-like proteolytic activities and degrades proteins in a processive manner, generating short peptides. The assembly and structure of the proteasome are dynamically controlled to enable the degradation of a wide variety of substrates and thereby regulate multiple cellular functions 19, 23, 24.
Ubiquitylation is often deregulated in many types of disease, including cancer, neurodegenerative, and immune disorders. For this reason, it is the focus of intense research that aims to develop effective inhibitors of UPS activity that could selectively kill altered cells. Bortezomib, also known as Velcade, is one of the best-characterized proteasome inhibitors and has been used to treat multiple myeloma, as well as a number of solid tumors 25, 26. Similarly, other proteasome subunits are current targets for drug development 27.
E3 ubiquitin ligase enzymes are responsible for the recognition of substrates for ubiquitylation. In humans, there are more than 600 E3 ligases, divided into three major families, the HECT, RING, and RING-between-RING (RBR) ligases 10, 28–31. Although they are highly selective enzymes, substrate recognition and ubiquitylation often depend on the cross-talk between different post-translational modifications. A well-characterized example is phosphorylation-dependent ubiquitylation, when phosphorylation is a direct pre-requisite for substrate recognition by the ubiquitylation machinery 32. However, phosphorylation can also block recognition of substrates, suggesting that phosphatases can also regulate this process 33.
Ubiquitylation of substrates followed by proteolytic degradation is a unidirectional process that involves the physical unfolding and cleavage of a protein. However, prior to being processed by the proteasome, ubiquitin removal can be catalyzed by DUBs, preventing proteasomal cleavage and resulting in protein stability. Five families of DUBs are known to exist, the majority of which are cysteine peptidases. DUBs are emerging as important players in development and the identification of a growing number of substrates has been the focus of much attention in a wide variety of systems. DUBs are implicated in chromatin regulation, transcriptional control, and the modulation of mitogenic pathways. Their function is often deregulated in malignancies, leading to a stabilization of oncogenic or anti-apoptotic factors 34, 35.
In all, ubiquitylation and protein degradation by the proteasome constitutes a highly regulated and evolutionarily conserved process that affects development and tissue physiology, and is frequently deregulated in disease. In the biology of stem cells, ubiquitylation plays key roles in self-renewal and cell fate specification. It provides an additional layer of stem cell regulation at the post-translational level and extends to multiple cellular processes, including the modulation of extracellular matrix composition, surface receptor trafficking and signaling, control of the cell cycle, transcription factor abundance, and deposition of epigenetic marks. Here, we discuss how ubiquitylation mediates the balance of cell fate decisions in different stem cell systems.
Ubiquitylation pathways in embryonic stem cells
Embryonic stem cells (ESCs) are derived from the inner cell mass of pre-implantation blastocysts 36, 37. They are pluripotent and have remarkable cellular plasticity, as they can differentiate to all somatic and germ cell lineages of the embryo proper. ESCs can be cultured in vitro maintaining their unique characteristics indefinitely. Additionally, they can differentiate to various cell types under proper culture conditions. These unique properties of ESCs—including their rapid proliferation—make them exceptionally valuable to study mechanisms that dictate protein stability. Ubiquitylation pathways can modulate ESC functions on multiple levels, and thus, there is increasing interest in their use for the discovery of drugs that alter protein abundance.
At the level of transcription, a core factor circuitry consisting of Oct4, Sox2, Nanog, and c-Myc regulates the balance between ESC self-renewal and differentiation. These proteins directly control the levels of each other, creating a positive feedback loop that sustains high levels of expression 38. Furthermore, they can form complex networks with multiple transcription and chromatin-regulating factors. The dynamic interactions among these components ultimately regulate differentiation and lineage specification processes 39. Changes in transcription factor abundance can result in the tipping of this balance toward specific patterns of differentiation. As a result, regulation at the post-translational level by ubiquitylation is crucial and can specify these processes.
There are several examples of ubiquitin-modulating enzymes that control transcription factor abundance in ESCs. The HECT family E3 ligase Wwp2 has been suggested to ubiquitylate Oct4 in both mouse and human ESCs 40, 41, decreasing Oct4 transcriptional activity and leading to its proteasomal degradation (Fig 1A). In the same studies, Wwp2 was also proposed to undergo auto-ubiquitylation, providing an additional level of complexity 42. Although there are a lot of open questions regarding this function in vivo, as well as how ubiquitylated Oct4 interacts with additional ESC components, these studies suggest that ubiquitylation fine-tunes self-renewal by controlling Oct4 levels and activity. As expression levels of Wwp2 correlate with pluripotency, it would be interesting to investigate how these functions control cellular reprogramming, given that Oct4 is a crucial factor in iPS generation 43.
Figure 1.
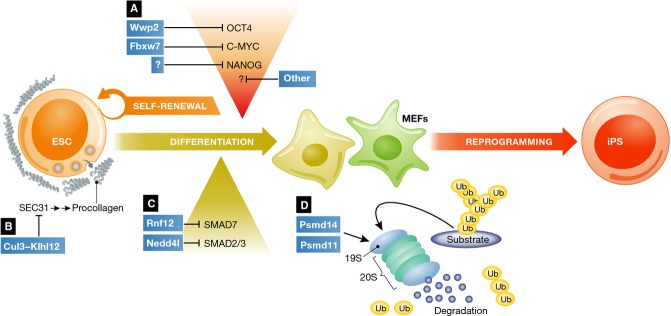
Ubiquitylation regulates ESC pluripotency, differentiation and iPS cell generation.
(A) The E3 ligases SCFFbxw7 and Wwp2 regulate core transcription factor—such as c-Myc and Oct4—abundance and functions in ESCs. Additional enzyme–substrate pairs may control transcriptional regulation of ESCs. SCFFbxw7 controls cellular reprogramming and iPS generation, in addition to differentiation, through c-Myc stabilization. (B) Cul3-Klhl12 ubiquitylates Sec31, regulating COPII vesicle size and procollagen export to the extracellular matrix. (C) Ubiquitylation regulates signaling components in ESCs. Nedd4l and Rnf12 regulate Smad2/3 and Smad7 levels, respectively. (D) Enzymatic functions of the proteasome control self-renewal and differentiation of ESCs, as well as cellular reprogramming. Psmd14 and Psmd11 regulate 19S regulatory particle activity and assembly, respectively.
Another example of transcription factor modulation in ESCs is c-Myc ubiquitylation by SCFFbxw7 44 (Fig 1A). This interplay illustrates how ubiquitylation orchestrates ESC differentiation. c-Myc is primed for ubiquitylation by Gsk3-dependent phosphorylation on Thr58, creating a phosphodegron that promotes its proteasomal degradation 45. Loss of c-Myc protein, a key transcriptional determinant of ESC function, induces an irreversible transition toward differentiation. Furthermore, degradation of c-Myc by SCFFbxw7 is involved in cellular reprogramming, as mouse embryo fibroblasts in which Fbxw7 is depleted are able to reprogram to iPS cells more efficiently 44. These results demonstrate the importance of dynamic ubiquitylation events on the establishment of cell fate specification.
Similar to Oct4 and c-Myc, the levels of Nanog are also regulated by ubiquitylation 46 (Fig 1A), and this process has been linked to phosphorylation 47. Nanog is known to be a phosphoprotein in mouse ESC 48, and recent efforts in human ESCs have tried to understand this post-translational cross-talk. Phosphorylation of Nanog alters its degradation rate, resulting in an increased stability in human ESCs 47. Its ubiquitylation and proteasomal degradation depend on specific sequence motifs, deletion of which has dramatic effects on Nanog stabilization 46. Although the E3 ligase responsible for this activity remains elusive, the maintenance of Nanog levels has a big impact on ESCs, as elevated Nanog expression sustains LIF-independent self-renewal 49. Therefore, it is intriguing to speculate that regulation at the post-translational level by ubiquitylation can have similar effects. However, additional work needs to clarify previous efforts to this direction. Interestingly, stemness proteins, such as Nanog, have been linked to malignancies and correlate with tumor progression and poor prognosis 50, 51. As the enzymes implicated in the post-translational control of Nanog levels have also been associated with pathology 52, a tempting possibility is that the ubiquitylation and proteasomal degradation of key pluripotency factors is not only a determinant of ESC differentiation but can also determine malignant transformation and disease progression.
In addition to level of transcription factors, ubiquitylation can regulate other aspects of ESC function, such as receptor signaling and vesicle trafficking. Ubiquitylation mechanisms have been elegantly linked to secretion and support of the extracellular matrix 53. The ubiquitin ligase Cul3—through its adaptor Klhl12—was found to monoubiquitylate Sec31, a basic constituent of COPII vesicles in ESCs (Fig 1B). This ubiquitylation controls the size of COPII vesicles, enabling the export of large vesicle cargo, such as procollagen fibers. These groundbreaking findings demonstrate that ubiquitin pathways control not only intracellular constituents, but also the extracellular space, expanding the perception of how ubiquitylation regulates integrin signaling and cell division in early embryo development.
Ubiquitylation also impacts signal transduction in ESCs, shaping their differentiation potential toward specified lineages. Components of the TGF-β signaling pathway, for example, are well-characterized targets of ubiquitylation. The E3 ligase Nedd4L can tag phosphorylated Smad2/3 for degradation in ESCs 54, skewing differentiation toward mesodermal lineages (Fig 1C). On the other hand, the inhibitory Smad7 was recently suggested to be a target of Rnf12 55. ESCs deficient of Rnf12 are unable to induce the formation of anterior mesoderm or inhibit neuronal differentiation when challenged with Activin and BMP/Smad, respectively.
Collectively, the above examples outline the diverse and complementary nature of ubiquitin-mediated modifications in ESCs. Relative protein abundance, vesicle trafficking and signal transduction, all regulated by ubiquitylation, control important cell fate decisions. Thus, the functional role of ubiquitin extends to fundamental stem cell processes, emphasizing its importance in development.
Finally, we would like to focus on the role of the proteasome in stem cells, as a machinery that balances opposing developmental processes. Interestingly, the various regulatory particles have different effects on proteasomal activity. As a result, their functions are distinct in ESCs. For instance, the 11S/PA28 activator is required to eliminate oxidatively damaged proteins during differentiation 56, 57. On the other hand, the 19S regulatory particle is essential for maintenance of self-renewal and turnover of ubiquitylated proteins. In mouse ESCs, the deubiquitylating enzyme Psmd14, an integral subunit of the 19S particle, is necessary for proper self-renewal 44 (Fig 1D). Loss of Psmd14 leads to differentiation with the simultaneous accumulation of polyubiquitylated proteins, which can be fully rescued by the restoration of this enzymatic activity. Furthermore, Psmd14 is required for iPS cell generation, further emphasizing the importance of proteasome function in pluripotency (Fig 1D). In human ESCs, PSMD11—which is a component of the proteasome lid—was shown to play an instrumental role in the assembly of the 20S core with the 19S particle, thus regulating proteasome activity 58. Decreased levels of this subunit lead to diminished cleavage activity and differentiation. Additional work is certainly needed in order to clarify the exact changes in proteasome architecture and identify the specific effects on stem cell elements that trigger differentiation. However, the above examples demonstrate the regulatory roles that proteostasis, and therefore the proteasome subunits, have on stem cell biology. In agreement with this, proteasomes have been suggested to restrict permissive transcription by degrading pre-initiation complexes at loci important for development, thereby preventing differentiation 59. Conversely, transcription factors such as OCT4 have been shown to directly or indirectly control the expression of multiple proteasome subunits in an interesting feedback loop 60. In summary, the work discussed above demonstrates the diversity and importance of ubiquitylation functions in ESC regulation.
Ubiquitylation pathways in adult stem cells
The regenerative potential of tissues relies on specialized subsets of multipotent stem cells that can give rise to all the cells that make up a given tissue. Those tissue-specific stem cells maintain a tightly controlled balance between quiescence, self-renewal, and differentiation. However, many of the molecular mechanisms governing stem cell fates are poorly understood. Some of them are common for several types of stem cells, whereas others are tissue specific. The ubiquitin system has an important role in the regulation of fundamental cellular functions such as cell cycle, DNA damage repair, protein quality control, and transcription; however, little is known about its impact in regulating adult stem cell differentiation and lineage specification. In the last years, the development of several E3 ligase-knockout mouse models has revealed important functions of the ubiquitin system in adult stem cell biology.
Hematopoietic stem cells as a paradigm of regulation by ubiquitin
Hematopoietic stem cells have the capacity to regenerate the blood cell lineage throughout the life of an organism 61–63. This capacity depends on their remarkable self-renewal and differentiation properties. Most of the HSCs remain quiescent in specialized niches of the bone marrow. Only in response to specific stimuli, HSCs can re-enter the cell cycle and self-renew or differentiate 64. Quiescence and self-renewal prevents exhaustion of the HSC, whereas differentiation induces the production of the different cell lineages. Therefore, the molecular mechanisms controlling the balance between quiescence, self-renewal, and differentiation are tightly regulated. Improper control of this equilibrium may result in hematopoietic failure or cancer 65. The regulation of these cellular events is achieved on two different levels. First, in the HSC niche, growth factors, chemokines, cytokines, and other secreted molecules constitute cell-extrinsic networks controlling HSC maintenance 66. Second, complex cell-intrinsic signaling pathways involving cell cycle, proliferation, growth, and survival have been shown to be essential in HSC homeostasis 65, 67–73. The UPS plays an important role in regulating protein functions required in both networks. Ubiquitylation of Notch 74 and c-Myc 68, 71, 72, 75, 76 are among the most characteristic examples of such regulation. Moreover, impaired ubiquitin-mediated regulation of HSCs has been linked to malignant transformation events 76.
E3 ligases controlling HSC quiescence
Loss of HSC quiescence leads to hematopoietic exhaustion, accumulation of replicative stress, and leukemia 77, 78. The impaired activity of some E3 ubiquitin ligases has been related to the loss of quiescence and expansion of the HSC compartment.
The RING E3 ligase c-Cbl negatively regulates Notch 79, c-Kit 80, and STAT5 81, 82, all of which are essential in HSC homeostasis (Fig 2A). c-Cbl-deficient mice have a cell-autonomous increase in HSCs 83. Similarly, c-Cbl-knockout HSCs show enhanced proliferation and reconstitution capacity in competitive bone marrow transplantations. The lack of this E3 ligase promotes self-renewal and aberrant proliferation of stem cells (Fig 2B). All these data support that c-Cbl maintains quiescence of the HSC compartment. Eventually, c-Cbl-knockout mice develop myeloid proliferative disorders and acute myeloid leukemia 84. Interestingly, CBL is mutated in 5–15% of human myeloid proliferative disorders 85–89. However, more studies are necessary to understand whether those mutations are drivers of leukemia.
Figure 2.
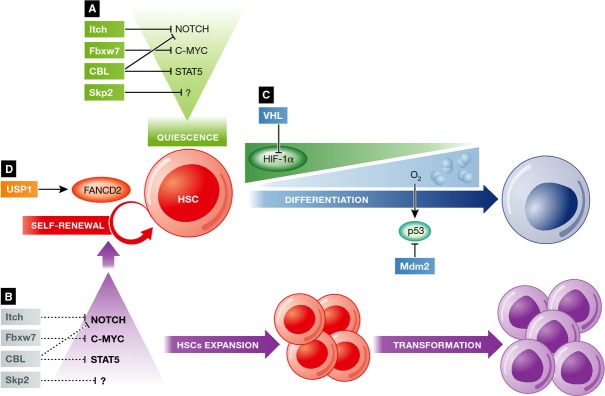
The ubiquitin-proteasome system maintains the balance between quiescence, self-renewal, and differentiation of HSCs.
(A) The E3 ligases Itch, SCFFbxw7,CBL and SCFSkp2 maintain the quiescence of the HSC by targeting Notch, c-Myc, and STAT5, among others. (B) Loss of any of those E3 ligases results in self-renewal factor upregulation. As a consequence, HSCs re-enter the cell cycle and divide. The aberrant expansion of HSCs results in hematopoietic exhaustion or leukemia. (C) HSCs need HIF-1α in order to survive in the hypoxic niche. However, during differentiation, they migrate out of the niche. The E3 ligases VHL and MDM2 play an important role in the adaptation to the new environment. HIF-1α must be degraded by VHL for migration and differentiation to occur. Additionally, as increase in ROS activates the p53 pathway, MDM2 controls the levels of p53, promoting cell survival. Impaired activity of USP1, VHL, or MDM2 alters HSC self-renewal and differentiation capacities, resulting in hematopoietic failure. (D) During HSC self-renewal, the DUB USP1 promotes the activation of FANCD2, which is essential for DNA damage repair upon replicative stress.
Similar to c-Cbl, the HECT E3 ligase Itch is a negative regulator of HSC self-renewal and proliferation 90. Itch-deficient mice show increased numbers of HSC and enhanced self-renewal. These phenotypes are attributed to deficits in the ubiquitylation of Notch (Fig 2A), a well-known target of Itch. Indeed, Notch downregulation in Itch-deficient mice can partially rescue the effects on HSCs 29. However, loss of Itch does not promote leukemia.
In contrast, the E3 ligase SCFFbxw7 is mutated in a significant portion of human tumors, including T-cell acute lymphoblastic leukemia (T-ALL) 91–93. This E3 ligase controls the stability of key hematopoietic regulators such as Notch 94, 95, c-Myc 96, 97, Cyclin E 98, 99, and Mcl-1 100, 101 (Fig 2A). Germline deletion of SCFFbxw7 results in embryonic lethality due to hematopoietic and vascular defects 95, 101. Conditional deletion of Fbxw7 in the hematopoietic system leads to constant HSC proliferation and eventually exhaustion of that population 74, 102 (Fig 2B). As a consequence, a significant percentage of Fbxw7-knockout mice develop anemia. Besides the increased proliferation of HSCs, these cells are unable to compete in bone marrow transplantation. Therefore, Fbxw7 deficiency results in a global loss of quiescence in the HSC compartment and consequently exhaustion of hematopoiesis. Accordingly, genes involved in HSC quiescence are downregulated in Fbxw7-deficient LSKs, which are the primitive hematopoietic cells that have the ability to self-renew. This may be due to changes in the stability or transcriptional activity of one or more of the Fbxw7 targets, such as c-Myc. Furthermore, some aged mice develop T-ALL due to the stabilization of c-Myc, which prevents T cells from exiting the cell cycle 74, 103. Interestingly, Fbxw7 was recently shown to modulate leukemia-initiating cells by the regulation of c-Myc stability 75.
Another important regulator of HSC self-renewal and quiescence is the E3 ligase SCFSkp2 (Fig 2A), which triggers ubiquitylation and degradation of cell cycle inhibitors such as p27 104. Upregulation of Skp2 has been related to increased proliferation and tumorigenesis 105. In contrast, depletion of Skp2 in long-term HSCs (LT-HSCs) can promote the loss of quiescence and proliferation 106, 107 (Fig 2B). While p21 and p27 levels are normal in the Skp2-null cells, Cyclin D1 is upregulated. However, Cyclin D1 is not a direct target of Skp2. Therefore, the molecular mechanism driving this proliferative phenotype in stem cells is still unknown.
E3 ligases in HSC differentiation
Similar to quiescence, proper control of HSC differentiation is essential for the regulation of hematopoiesis. One of the hallmark characteristics of the HSC niche is its low-oxygen tension, essential for stem cell quiescence and functions 108. In order to adapt to this hypoxic niche, HSCs have developed appropriate molecular mechanisms for stem cell survival. HIF-1a is the master regulator that orchestrates this hypoxic transcriptional program 109. When HSCs differentiate, they move out from the HSC niche and HIF-1a is degraded through the E3 ligase VHL 109 (Fig 2C). Indeed, VHL plays an essential role in the exit from quiescence and HSC differentiation. Loss of one or two alleles of VHL induces HSC quiescence, determined by both an increase in the total numbers of LSK cells, which contain the HSCs population, and in the numbers of LSKs in G0. As a consequence, there is attenuated differentiation of cells in the peripheral blood. Therefore, VHL is essential for exit from quiescence and the initiation of HSC differentiation.
In addition to the above example, the hypoxic bone marrow niche keeps low levels of reactive oxygen species (ROS). As stem cells move out from the niche to proliferate and differentiate, the concentration of ROS increases. Given that ROS are DNA-damaging agents, cells activate p53 pathways. The RING E3 ligase MDM2 regulates p53 stability, facilitating the survival of HSCs upon those microenvironmental changes. The ablation of MDM2 in the hematopoietic system leads to the stabilization of p53, resulting in cell cycle arrest, senescence, and apoptosis of HSC and progenitor cells 110 (Fig 2C). Several studies have demonstrated that this is a cell-intrinsic effect alleviated by p53 downregulation or treatment with antioxidants 111, 112. Therefore, MDM2 is required to regulate p53 levels as ROS levels increase when the HSCs migrate away from the niche.
In the adult organism, most HSCs remain quiescent and are thus protected from the DNA damage inherent to DNA replication and from molecular species resulting from cellular metabolism. However, as HSCs re-enter the cell cycle to divide and differentiate, they are exposed to such stress. An important DNA repair mechanism is the Fanconi anemia pathway. Monoubiquitylated FANCD2 is recruited to chromatin to mediate this process 113, and the cysteine protease USP1 deubiquitylates FANCD2 (Fanconi anemia D2) to end this function 114 (Fig 2D). Loss of Usp1 causes detrimental phenotypes in mice 115, 116, similar to those observed in patients deficient for any of the Fanconi anemia pathway proteins. These include aplastic anemia, developmental abnormalities, and increased cancer susceptibility due to increased genomic instability 117. Accumulation of DNA damage and apoptosis in the HSC compartment explains bone marrow failure phenotypes in those patients. As a result, the deubiquitylating activity of Usp1 demonstrates its key role in HSC protection against DNA damage. However, it is unclear whether this is only through its regulation of FANCD2 or other targets are also implicated.
Ubiquitylation in epidermal stem cells
The skin is a multilayer organ that protects organisms against external aggressions. It constitutes one of the tissues with the best-characterized hierarchical organization. Stem cells are found in the basal layer of the epidermis and generate several types of progenitors ensuring the high turnover rate of the epithelium. In addition, the bulge region of the hair follicle in mice contains a population of multipotent stem cells that can give rise to all epithelial cell lineages within hair follicles during normal hair growth 118. Surprisingly, non-hair follicle stem cells can contribute to the formation of hair follicles in response to wounding 119, demonstrating the dynamic characteristics of this system. Although only some of those stem cells remain quiescent, most of the epidermal stem cells can divide symmetrically or asymmetrically in order to self-renew or differentiate. This balance between quiescence, symmetric, or asymmetric cell division is controlled by a number cell-autonomous molecular mechanisms and interactions between stem cells and their microenvironment 120. As in other tissues, disequilibrium between self-renewal and differentiation results in pathologies like cancer 121.
The canonical Wnt/β-catenin signaling pathway has been implicated in maintaining stem cell homeostasis in epithelial tissues such as the skin, mammary gland, and intestine. Furthermore, deregulation of this pathway often leads to the generation of epithelial cancers 122. In the absence of active Wnt signaling, intracellular β-catenin is phosphorylated and targeted for proteolytic degradation. High levels of β-catenin lead to hair growth and ectopic hair follicle formation in transgenic mice 123, whereas inhibiting β-catenin blocks the formation of hair during development 124. In addition, transplant experiments in pre-cancerous murine skin cancer stem cells show that β-catenin signaling is essential for tumorigenesis 118. Interestingly, the E3 ligase Smurf2 targets Smad7-bound β-catenin for degradation 125 (Fig 3A). Transgenic mice overexpressing Smad7 show reduced levels of β-catenin and Wnt signaling inhibition, resulting in the delay of hair development. Under physiological conditions, the levels of Smad7 in keratinocytes are low; however, its knockdown leads to overexpression of β-catenin and Wnt signaling. This suggests that low levels of Smad7 are required in stem cells to maintain a proper balance of β-catenin/Wnt signaling. Accordingly, whether the overexpression of Smad7 in β-catenin-dependent cancer stem cells could abolish tumorigenesis is an intriguing possibility that awaits testing.
Figure 3.
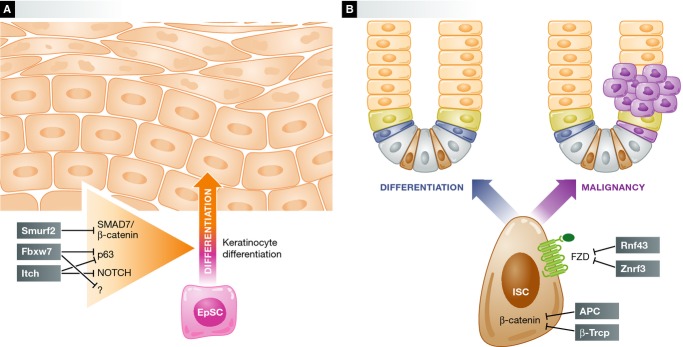
Ubiquitylation maintains the equilibrium between differentiation and transformation in adult stem cells.
(A) The E3 ligases Itch, SCFFbxw7, and Smurf2 regulate levels of Notch, p63, and β-catenin in epidermal stem cells, mediating keratinocyte differentiation. (B) In intestinal stem cells, the Wnt signaling pathway is modulated by ubiquitylation. Rnf43 and Znrf3 control Frizzled proteins, whereas APC and SCFβTrcp regulate β-catenin degradation, collectively controlling differentiation in the intestine. The alteration of this balance due to mutation results in tumor formation.
Another example of ubiquitin-mediated regulation is that of the HECT E3 ligase Itch, which similarly to its function in the hematopoietic system, has important roles in epidermal stem cells. Itch was originally identified through genetic studies aimed to examine the agouti locus. The 18H mutation associated with a darker color arises from an inversion disrupting the agouti and Itch genes 126. Interestingly, some Itch substrates such us p63, Notch, Gli1, c-Jun, JunB, Erb4 are transcription factors controlling epidermal stem cell maintenance and keratinocyte differentiation 127, 128–130 (Fig 3A). Itch-deficient mice develop severe immune deregulation 131 and a thickening of the epidermis 132. In skin epithelia, the p63 isoform NΔp63α supports the proliferative potential of basal cells. Moreover, its downregulation is required during keratinocyte differentiation 133. Aberrant accumulation of NΔp63α and Notch during keratinocyte differentiation might partially explain the epidermal hyperproliferative phenotype of Itch-null mice. However, the precise role of Itch in epidermal stem cells remains unknown. Furthermore, SCFFbxw7 has been proposed to target p63 during keratinocyte differentiation and DNA damage regulation 134 (Fig 3A). As in other stem cells, SCFFbxw7 could regulate the proliferation of epidermal stem cells.
Ubiquitylation in intestinal stem cells
In the mammalian intestine, rapidly dividing stem cells are confined to the crypts of Lieberkuhn. They continuously self-renew and differentiate to regenerate all tissues every 4–5 days. Alterations in these stem cell functions might result in the expansion of the stem-like cells, adenomas, and cancer 135. As in skin, Wnt signaling was one of the first mechanisms implicated in the control of gut stem cells by maintaining active cell divisions 136.
Canonical activation of Wnt signaling leads to the stabilization of β-catenin 136. In the absence of Wnt ligand, β-catenin is sequestered in a multiprotein degradation complex containing the scaffold protein Axin, the tumor suppressor adenomatous polyposis coli APC (Fig 3B) and the kinases CKI and GSK3β. Upon sequential phosphorylation at serine and threonine residues, β-catenin is ubiquitylated by the E3 ligase SCFβ-TrCP and subsequently degraded by the proteasome 137 (Fig 3B). Mutations in APC or β-TrCP result in β-catenin stabilization and nuclear translocation, constitutively activating its transcriptional targets, which induces adenoma formation and colon cancer 138, 139 (Fig 3B). Although mutations in β-TrCP have been only found in gastric and endometrial cancer, transgenic mice overexpressing either wild-type or loss-of-function β-TrCP develop tumors in a wide variety of organs through β-catenin activation 140, 141. For these reasons, β-TrCP may be important during stem cell differentiation.
However, β-TrCP is not the only mediator of Wnt signaling. In a recent landmark study, the RING-family transmembrane E3 ligases RNF43 and ZNRF3 were shown to modulate the Wnt pathway, regulating the functions of the LGR5-positive crypt stem cells 142 (Fig 3B). According to this study, RNF43 and ZNRF3 target frizzled receptors for degradation. Deletion of both genes in mouse intestinal epithelium induces rapidly growing adenomas due to an expansion of the LGR5-positive stem cells. Inhibition of Wnt secretion decreased the proliferation of organoids derived from those adenomas 142. This groundbreaking work provides an excellent example of signaling cascade modulation by ubiquitylation in stem cell function.
Another E3 ligase that is expressed in the nucleus of the crypt stem cells is SCFFbxw7. Loss of Fbxw7 alters intestinal epithelium homeostasis and induces the development of adenomas 143. In the context of APC deficiency, ablation of Fbxw7 accelerates intestinal tumorigenesis, promoting an accumulation of β-catenin in adenomas that normally have a long latency. Intestinal alterations and a susceptibility to adenoma formation suggest that lack of Fbxw7 results in the expansion of stem cell crypts. However, further analysis is required to validate the role of Fbxw7 in the intestinal stem cells.
Ubiquitylation in neural stem cells
Multipotent neural stem cells (NSCs) exist both in embryonic and in adult tissues of the nervous system. They reside in various distinct anatomical sites and are able to maintain and specify neurogenesis during early development, as well as throughout post-natal life. Among various signaling pathways that regulate NSC maintenance, differentiation and specification, the Notch pathway is of crucial importance. Notch can inhibit neurogenesis and maintain glial progenitor cells 144, 145. In this stem cell system, ubiquitin-mediated regulation plays an important role. For instance, the RING E3 ligase Mbi1 shapes Notch regulation by ubiquitylating Notch ligands in neighboring cells. Loss of Mbi1 results in aberrant Notch activation, which leads to premature neurogenesis 146. The perturbation of Notch ligand ubiquitylation in the absence of Mbi1 results in their defective endocytosis, sustaining Notch activation 147. This important function places Mbi1 as an upstream regulator of Notch signaling.
The self-renewal of NSCs must be also regulated by suppression of neural differentiation genes. The transcription factor REST is an important repressor of differentiation toward neurons and constitutes another example of ubiquitin-mediated cell specification. SCFβ-Trcp is the E3 ligase that mediates REST proteasomal degradation 148. Knockdown of β-Trcp inhibits neuronal differentiation from ESCs. Similarly, β-Trcp expression levels are upregulated during the induction of neuronal differentiation.
Conversely, NSC differentiation must be accompanied by the loss of factors that promote self-renewal. N-myc is an essential transcription factor that promotes cycling and controls the transcriptional program of NSCs. The HECT-type E3 ligase Huwe1 was shown to target N-myc for polyubiquitylation and proteasomal degradation, allowing proper neuronal differentiation 149. Huwe1-knockout ESCs fail to differentiate toward neurons in vitro and, similarly, depletion of Huwe1 in vivo leads to improper maintenance of stemness characteristics in the mouse brain.
Beyond the described functions in neural development, E3 ligases are often mutated in neurodegenerative diseases. The mouse model of the RING E3 ligase Listerin is an example of the importance of proper ubiquitylation in human disease 150. Mutant mice for the lister gene present defective neuronal and motor functions. Axons and neurons are degenerated, leading to muscle atrophy. Another example of E3 ligases implicated in neurodegenerative disease is the RING ligase Parkin. Mutations in the Park2 locus, which encodes Parkin, were identified in Parkinson's disease 151. Parkin was found to bind the E2-conjugating enzyme UbcH8, promoting the ubiquitylation and degradation of the synaptic vesicle-associated protein CDCrel-1 152. Several other substrates have been proposed and loss-of-function Parkin mutations are associated with a loss of dopamine neurons 153.
Histone ubiquitylation in stem cell function and disease
Although ubiquitylation often leads to the formation of polyubiquitin chains that can ultimately lead to proteasomal degradation, other types of ubiquitin conjugation to substrate proteins do not mediate proteolysis. Examples of such modifications are monoubiquitylation and conjugation of polyubiquitin chains linked through lysines other than K48. These play a pivotal role in stem cell biology, as they can regulate a plethora of different processes, such as regulation of histone function and gene expression, as well as receptor endocytosis and DNA repair 16.
Stem cells impose plasticity in chromatin structure and dynamics, processes that facilitate the rapid establishment of different gene expression patterns after differentiation stimuli. Nucleosomes constitute the basic units of chromatin and are composed of 147-bp DNA fragments surrounding octamers of histones H2A, H2B, H3, and H4, which are present in dimers. The post-translational modification of histone tails can result in alteration of histone physical properties, leading to alterations in protein compaction. In addition, they can serve as a platform for the recruitment of transcription factors, enzymes, and other chromatin-associated proteins 154. Histone ubiquitylation thus plays substantial roles in the regulation of gene expression programs during stem cell self-renewal or differentiation, mediating transitions in chromatin architecture and organization 155.
As opposed to other modifications such as methylation, phosphorylation, or acetylation that are modest in size, histone monoubiquitylation covalently links a larger approximately 8-kDa protein to histones, affecting chromatin structure. In mammals, H2B-monoubiquitylation (H2B-Ub) on Lys120 is catalyzed by the E3 ligases RNF20 and RNF40 156, which form a complex, and the E2-conjugating enzymes hRad6A and hRad6B 157 or UbcH6 158, 159 (Fig 4A). Chromatin immunoprecipitation (ChIP) studies showed that H2B-Ub is closely associated with transcriptionally active regions in chromatin and RNA polymerase II elongation 160–162. Given that it constitutes a bulky modification associated with open chromatin structure, it was postulated that it causes steric effects to allow the opening of chromatin conformation 163. H2B-ub has been also shown to regulate nucleosome assembly and disassembly 164. This is achieved during transcription elongation through the histone chaperone FACT, which can toggle the deposition of H2B-ub. FACT can modulate H2A/H2B removal to facilitate elongation by RNA polymerase II, and it can restore nucleosome assembly, promoting chromatin dynamics and configuration during transcription 159. The Paf1 transcriptional elongation complex is also involved in H2B-Ub deposition, as it plays an active role in the recruitment of RNF20 to RNA polymerase II among other functions 165.
Figure 4.
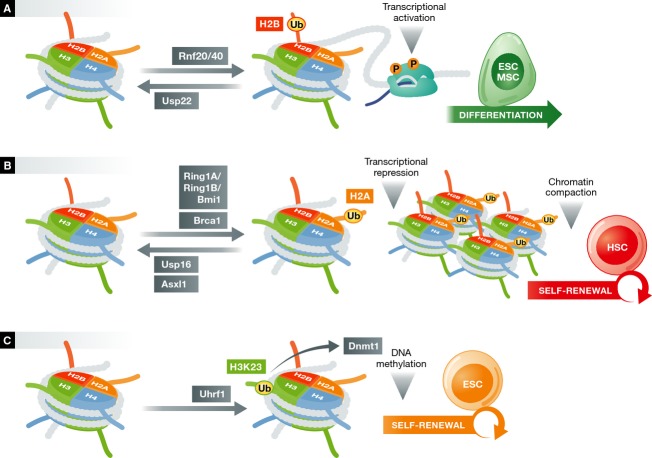
Regulation of chromatin functions in stem cells by ubiquitylation.
(A) H2B ubiquitylation marks open chromatin and facilitates transcription elongation by RNA polymerase II. The E3 ligases RNF20 and RNF 40, and the DUB USP22, control H2B-Ub deposition and transcriptional activation, resulting in ESC and MSC differentiation. (B) H2A ubiquitylation is deposited by either the PRC1 complex, which consists of RING1A/RING1B/BMI1- or the E3 ligase BRCA1, and is associated with transcriptional repression and chromatin compaction. The DUBs USP16 and Asxl1 can reverse this process, regulating the functions of hematopoietic and mammary stem cells. (C) H3K23 ubiquitylation is catalyzed by the E3 ligase Uhrf1. This recruits Dnmt1, which maintains DNA methylation at sites of replication.
Given that H2B-Ub modification can have marked effects in gene expression, it plays an important role in stem cell differentiation and adaptation of transcriptional programs. H2B-Ub is significantly upregulated during differentiation of human mesenchymal stem cells (hMSCs) 166 (Fig 4B). Indeed, hMSC differentiation is inhibited upon the depletion of RNF40, leading to significant changes in the transcriptional programs of osteoblast and adipocyte lineages. Similarly, H2B-Ub is required for the optimal differentiation of embryonic stem cells, and depletion of RNF20 is associated with impaired induction of differentiation-associated gene subsets 167. However, the opposite is observed during myoblast differentiation, where deposition of this histone mark is rapidly downregulated, giving rise to myotube formation 168. Another direct implication of H2B-Ub in gene expression and its implication in cellular identity and morphogenesis is its requirement for the transcriptional activity of Hox genes. Importantly, H2B-Ub was the first histone mark directly implicated in the modification of another histone in a cross-talk mechanism. H2B-Ub is required for H3K4me3 and H3K79me3 formation by complexes containing Set1- and Dot1-histone methyltransferases, respectively 169. Consistent with that, knockdown of RNF20 leads to a decrease in not only H2B-Ub, but also H3K4 and H3K79 methylation 156.
Besides its positive roles in RNA polymerase II elongation and chromatin compaction, H2B-Ub has also been linked to transcriptional repression. RNF20 is associated with repression of the proto-oncogenes c-Fos and c-Myc 161. In addition, in RNF20-depleted cells, epidermal growth factor (EGF)-induced genes are de-repressed, suggesting that H2B-ub can have tumor suppressor activity. Similar to E3 ligases, DUBs specific for H2B-Ub can also regulate transcriptional activity and chromatin functions. USP22 is the major H2B-Ub DUB in mammalian cells (Fig 4A). It has been shown to deubiquitylate H2B on promoters, regulating androgen and estrogen receptor-mediated transcription 170. Furthermore, USP22 is implicated in the activation of c-myc targets in tumor-initiating stem cells 171 and is correlated with tumor metastasis and poor patient prognosis 172, 173. In Drosophila, the H2B-Ub DUB Scrawny (scny) is involved in germline, epithelial, and intestinal stem cell maintenance 174. Aberrant Notch pathway signaling was observed in scny Drosophila mutants, suggesting that H2B-Ub functions through the silencing of genes required for differentiation.
Another important mechanism regulating stem cell functions involves H2A ubiquitylation on Lys119. The Ring1B component of the polycomb repressive complex 1 (PRC1) was shown to be the main E3 ligase for this enzymatic function in mammalian cells (Fig 4B), as loss of Ring1b dramatically decreases H2A-ub deposition globally 175–177. In contrast to H2B-Ub, H2A-Ub plays a repressive role in transcription 175. In ESCs, it has been associated with poised RNA polymerase II at bivalent genes 178. Additionally, H2A-Ub correlates with the loss of occupancy of RNA polymerase II phosphorylated at Ser5 and Ser2, which mark initiation and elongation 158. During development, H2A-Ub controls Hox gene silencing 176, 179 and X-chromosome inactivation 34, 177, among other functions. Other RING-finger proteins of the PRC1 complex, Ring1A and Bmi1, stimulate H2A-Ub deposition 176, 180, suggesting that Ring1A/Ring1B/Bmi1 act in concert in order to modulate H2A functions. In support of these findings, Bmi1 was shown to be essential for hematopoiesis, as bone marrow HSCs lacking Bmi1 are unable to self-renew properly 69, 70 (Fig 4B). Bmi1 is also overexpressed or amplified in many leukemias 181. Similar to HSCs, Bmi1 is known to regulate neural and mammary stem cell self-renewal, preventing differentiation 182–184.
Besides ubiquitylation by the PRC1 complex, other E3 ligases have been proposed to act on H2A to regulate its functions. BRCA1 is a RING-finger E3 ligase with H2A-ub activity 185 (Fig 4B), which is stimulated by its association with BARD1, a RING-finger protein that lacks enzymatic activity 186. Recent work has associated BRCA1 with satellite regions of the genome 187, suggesting that its loss disrupts constitutive heterochromatin and leads to the deregulation of gene silencing, which suggests a tumor suppressor role. Loss of BRCA1 is implicated in multiple breast and ovarian cancers 188. Additionally, BRCA1 mutations are present in expanded luminal progenitor populations 189, consistent with proposed perturbations in their differentiation program.
In addition to E3 ligases, H2A-specific DUBs are also implicated in the regulation of gene expression during development. USP16 was demonstrated to mediate Xenopus embryo patterning through Hox gene silencing 190 (Fig 4B). Furthermore, it has been implicated in de-repression of transcription following DNA damage 191. In addition to USP16, the polycomb protein BAP1 has ubiquitin C-terminal hydrolase activity for H2A-Ub 192. In Drosophila, loss of BAP1 results in diminished deubiquitylation of H2A-Ub and aberrant Hox gene de-repression. In addition, its binding partner ASX, also a polycomb protein, is required for H2A-Ub DUB activity. Its mammalian counterpart, Asxl1, is often deleted or mutated in a number of hematologic malignancies, and Asxl1-knockout stem and progenitor cells present impaired self-renewal 193, 194 (Fig 4B). In addition to monoubiquitylation, H2A and its variant H2AX can be polyubiquitylated with K63-linked chains by the E3 ligases Rnf8 and Rnf168 195, 196. This occurs at foci of DNA damage and might serve as a platform in the recruitment of DNA repair-associated proteins.
In addition to H2A and H2B, the other core nucleosome histones, H3, and H4, as well as the linker histone H1, have been proposed to be regulated by ubiquitylation 197, 198. However, the relevance of most of these modifications in stem cell function remains unclear. Ubiquitylation of H3 at Lys23 (H3K23-Ub) by the RING-finger protein Uhrf1 was recently shown to modulate DNA methylation and DNA replication 199 (Fig 4C). The role of Uhrf1 was known to involve the recruitment of Dnmt1 to sites of hemi-methylated DNA 200, 201 to maintain DNA methylation in mammalian cells 202. A RING-finger mutant of Uhrf1 expressed in ESCs failed to maintain DNA methylation at DNA replication sites, suggesting a direct implication of H3K23-Ub in proper ESC differentiation.
Collectively, all the above mechanisms of regulation accentuate the importance of proteolytic-independent functions of ubiquitylation in stem and progenitor cell functions.
Ubiquitin regulation in cancer stem cells: potential therapeutic targets
Cancer stem cells (CSCs) are defined as a population that have the capacity to self-renew, differentiate, and regenerate the cells that originated the tumor. The concept of CSCs remains controversial and probably the role that CSCs play in tumor biology depends on individual types of malignancy. However, emerging evidence suggests that cancers consist of heterogeneous populations in which cancer-initiating cells can regenerate the bulk of the tumor 203. Therefore, the concept of CSCs will undoubtedly help us understand tumor biology and design novel therapeutic strategies.
As explained for different stem cell systems above, ubiquitylation controls self-renewal or differentiation through the regulation of different substrates. Acquisition of self-renewal capacity is one of the first tumorigenic events in most tumors. In fact, alterations of the ubiquitin pathway are known to promote tumorigenesis in mouse models by inducing stem cell-like phenotypes 29, 204, 205. One of the most studied examples is the E3 ligase SCFFbxw7. As discussed above, Fbxw7 can target several oncoproteins, such us Notch, c-Myc, and Cyclin E. Thus, it is not surprising that Fbxw7 acts as a tumor suppressor. In fact, Fbxw7 is located on chromosomal region 4q32, which is frequently lost in tumors. Moreover, Fbxw7 is mutated in several malignancies, such as cholangiocarcinoma, T-ALL, colon, endometrium, and stomach cancers 91, 99, 206, 207. Ablation of Fbxw7 in the hematopoietic compartment results in T-ALL, whereas deletion in the intestine promotes the development of adenomas. The upregulation of the Notch pathway in both systems explains the acquisition of self-renewal capacity in the transformed cells. Moreover, in the hematopoietic compartment, stabilization of c-Myc in Fbxw7 mutant mouse models has been shown to promote cancer-initiating cell populations 75 Interestingly, treatment of those mice with c-Myc inhibitors leads to T-ALL remission.
Sidebar B: Quantitative ubiquitin proteomics to study dynamic changes of stem cell identity.
In order to delve into the systemic details of post-translational regulation of cellular plasticity, specific substrate–enzyme interactions have been interrogated in the context of stem cell differentiation or cellular transformation. A lot of these studies have been performed primarily using epitope tagging and affinity-based approaches and have been very informative about the nature and specific details of the enzymatic functions orchestrating these processes 218. However, advances in the field of mass spectrometry in recent years enabled the use of proteome-wide studies in order to understand the dynamics of signaling networks, providing a holistic overview of cellular identity changes 219.
These powerful proteomic tools have been utilized in the past to study phosphorylation, acetylation or other small protein modifications in a system-wide manner 220. However, ubiquitylation poses additional challenges in comparison, considering its bulk size and chain branching. Nevertheless, a novel tool that has revolutionized this field is the use of monoclonal antibodies that recognize di-glycine moieties linked by an isopeptide bond to lysine side chains of proteins 221. These epitopes, which constitute the remnants of ubiquitylation events after tryptic digestion, can be biochemically isolated and subjected to mass spectrometry in order to identify whole-proteome ubiquitin signatures, while providing site-specific information 222.
Importantly, this methodology can be effectively combined with SILAC strategies, which allow for metabolic labeling of proteins in culture using isotopic amino acid variants 223. This powerful approach and methodology allows the detailed study of changes in ubiquitin conjugation in a quantitative manner in response to different stimuli. Such technology has been particularly useful to delineate the temporal changes of ubiquitylation in stem cell differentiation and reprogramming, allowing the correlation of relative protein abundance of self-renewal factors with differentiation timing 44. Additionally, it provides valuable information with regard to dynamics in site specificity and protein turnover after differentiation stimuli. Additional examples of this application include the identification of substrates for Cullin-RING ligases (CRLs) 224. Taken together, the applications described above can expand our knowledge on missing E3 ligase-substrate pairs and also on protein network dynamics during stem cell maintenance or differentiation.
Furthermore, the important roles of ubiquitylation in stem cell and cancer stem functions illustrate the importance of its machinery as therapeutic targets. The best example in cancer therapy is Bortezomib, or Velcade, which inhibits protein degradation by the proteasome and is approved in the United States for the treatment of relapsed multiple myeloma and mantle cell lymphoma. However, as Bortezomib affects all ubiquitin-tagged proteins, therefore lacking substrate specificity, it may cause undesired effects. In contrast, inhibition of one E3 ligase would lead to the accumulation of only a small number of substrates. Efforts to identify drugable cancer-specific targets include screening for altered requirements of ubiquitylation between stem and cancer stem cells. For instance, the comparison of primary glioblastoma cancer stem cells with neural stem cells revealed 28 E3 ligases whose downregulation promotes CSC differentiation or apoptosis 208. A model enzyme for drug targeting is MDM2, which controls p53 degradation, and it is associated with several malignancies. Cis-imidazolines and spirooxindoles can disrupt the interaction between p53 and MDM2, and these compounds are currently in clinical trials to explore their activity against human tumors 209. Additional efforts include in silico screens to identify compounds that inhibit SKP2 210. Inhibiting SCFSKP2 E3 ligase activity impairs the proliferation of a wide range of cancer cell lines. Additionally, it diminishes the prostate cancer stem cell properties of PC3 cells. Collectively, the above studies stress the importance of ubiquitylation in tumor-initiating cell biology. Manipulation of ubiquitin-regulating mechanisms can alter the oncogenic properties of cancer stem cells, leading to effective therapies.
Sidebar C: In need of answers.
As only few enzyme–substrate pairs have been characterized in stem cell functions, which additional E3 ligases and DUBs control differentiation or self-renewal in various stem cell systems?
How can technical advances in the field of proteomics help us understand ubiquitylation events in small stem cell populations in vivo?
How can we develop small-molecule inhibitors for specific components of the ubiquitin pathway? Given that enzymes often regulate multiple substrates in different cellular contexts, how can these strategies change the properties of specific stem cell systems?
Stem cells often represent < 1% of the total cell population in adult tissues. Can we identify ubiquitylation events that regulate the expansion of stem cell populations?
Can we identify “ubiquitin codes” in stem cells? Can different ubiquitin linkages and branching decipher alternative outcomes in the regulation of quiescence or differentiation?
Which enzymes differentially control tumor initiation, progression and relapse? As the field needs additional in vivo proof that ubiquitin pathways are important for these functions, what is the distinction between substrates in malignancies?
Which additional non-proteolytic functions of ubiquitylation result in alterations in membrane receptor repertoire, localization of factors and chromatin landscape during stem cell differentiation? How are these processes deregulated in cancer stem cells?
Conclusions and future directions
Since its identification, the UPS has emerged as an important regulator of different processes through the control of protein stability. Additionally, the identification of proteolytic-independent functions has linked the ubiquitin system to the regulation of signaling networks and epigenetic mechanisms. Together, the proteasome-dependent and independent functions of the ubiquitin system play an important role in stem cell quiescence, self-renewal, and differentiation. Only a few of the numerous E3 ligases and DUBs encoded in the human genome are well characterized. In addition, little is known about how others regulate stem cell fates. Therefore, the identification of the role and targets of additional enzymes could help us understand additional characteristics of stem cell biology. Moreover, identifying the mechanisms of deregulation of specific enzymes that impinge on tumor initiation and progression could lead to effective therapies against additional types of cancer. As this fast-moving field stands at the crossroads of proteomics, stem cell biology, and therapeutics, there are increasing expectations for effective manipulation of cellular systems and the discovery of new concepts in protein modification in cell plasticity.
“Ubiquitylation: mechanism and functions” Review series.
Previous issues of EMBO reports include:
Building and remodeling Cullin-RING E3 ubiquitin ligases, by Wade Harper et al
Ubiquitin in the immune system, by Henning Walczak et al
RBR E3 ligases at work, by Judith Smit & Titia Sixma
Dynamic survey of mitochondria by ubiquitin, by Mafalda Escobar-Henriques and Thomas Langer
Other reviews in this series, which will be published in consecutive issues of EMBO reports, will cover:
Understanding ubiquitylation one structure at a time, by Ronald Hay et al
Acknowledgments
We would like to thank all members of the Aifantis laboratory for useful discussions. I.A. is an investigator of the Howard Hughes Medical Institute and is receiving funds from National Institutes of Health (RO1CA133379, RO1CA105129, RO1CA149655, 5RO1CA173636) and the NYSTEM program of the New York State Health Department (NYSTEM-N11G-255). A.S. is supported by the NYSTEM institutional NYU Stem Cell Training Grant (C026880). M.G-R. is supported by the Ramon Areces Foundation. A.S., M.G., and I.A. wrote the review. A.S. and M.G-R. prepared the figures.
Glossary
- APC
adenomatous polyposis coli
- BMP
bone morphogenetic protein
- CKI
casein kinase 1
- CRL
Cullin-RING-ligase
- Dnmt1
DNA methyltransferase 1
- DUB
deubiquitylating enzyme
- FANCD2
Fanconi anemia group D2 protein
- Fbxw7
F-box/WD repeat containing protein 7
- GSK3β
glycogen synthase kinase 3 beta
- HECT
homologous to E3-AP C-terminus family of E3 ligases
- HIF-1a
hypoxia-inducible factor 1, alpha subunit
- iPS
induced pluripotent stem cell
- JAK
Janus kinase
- Lgr5
leucine-rich repeat containing G-protein-coupled receptor 5
- LSK
lineage-negative Sca1-positive cKit-positive cells containing the hematopoietic population
- Mbi1
mind-bomb 1
- Mdm2
mouse double minute 2 homolog
- Psmd
proteasome 26S subunit, non-ATPase
- REST
RE1-silencing transcription factor
- RING
really interesting new gene family of E3 ligases
- ROS
reactive oxygen species
- SILAC
stable isotope labeling by amino acids in culture
- Skp2
S-phase kinase-associated protein 2
- Smad
small body size/mothers against decapentaplegic; TGF-β signaling transcription factors
- STAT
signal transducer and activator of transcription
- T-ALL
T-cell acute lymphoblastic leukemia
- TGF-β
transforming growth factor beta
- UPS
ubiquitin-proteasome system
- USP1
ubiquitin-specific peptidase 1
- VHL
Von Hippel-Lindau
- Wnt
homologues of the Drosophila “wingless” signaling proteins
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Fuchs E, Chen T. A matter of life and death: self-renewal in stem cells. EMBO Rep. 2013;14:39–48. doi: 10.1038/embor.2012.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 3.Passegue E, Wagers AJ. Regulating quiescence: new insights into hematopoietic stem cell biology. Dev Cell. 2006;10:415–417. doi: 10.1016/j.devcel.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 6.Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 8.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 9.Catic A, Ploegh HL. Ubiquitin-conserved protein or selfish gene? Trends Biochem Sci. 2005;30:600–604. doi: 10.1016/j.tibs.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 11.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 12.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 15.Bedford L, Paine S, Sheppard PW, Mayer RJ, Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010;20:391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadowski M, Suryadinata R, Tan AR, Roesley SN, Sarcevic B. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 2012;64:136–142. doi: 10.1002/iub.589. [DOI] [PubMed] [Google Scholar]
- 17.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 18.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marques AJ, Palanimurugan R, Matias AC, Ramos PC, Dohmen RJ. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 20.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 22.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorokin AV, Kim ER, Ovchinnikov LP. Proteasome system of protein degradation and processing. Biochemistry (Mosc) 2009;74:1411–1442. doi: 10.1134/s000629790913001x. [DOI] [PubMed] [Google Scholar]
- 24.Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482:186–191. doi: 10.1038/nature10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caravita T, de Fabritiis P, Palumbo A, Amadori S, Boccadoro M. Bortezomib: efficacy comparisons in solid tumors and hematologic malignancies. Nat Clin Pract Oncol. 2006;3:374–387. doi: 10.1038/ncponc0555. [DOI] [PubMed] [Google Scholar]
- 26.Driscoll JJ, Woodle ES. Targeting the ubiquitin+proteasome system in solid tumors. Semin Hematol. 2012;49:277–283. doi: 10.1053/j.seminhematol.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 27.Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, et al. A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell. 2013;24:791–805. doi: 10.1016/j.ccr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125:531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8:209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smit JJ, Sixma TK. RBR E3-ligases at work. EMBO Rep. 2014;15:142–154. doi: 10.1002/embr.201338166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lau AW, Fukushima H, Wei W. The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis. Front Biosci (Landmark Ed) 2012;17:2197–2212. doi: 10.2741/4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi M, Duan S, Jeong YT, Horn M, Saraf A, Florens L, Washburn MP, Antebi A, Pagano M. Regulation of the CRL4(Cdt2) ubiquitin ligase and cell-cycle exit by the SCF(Fbxo11) ubiquitin ligase. Mol Cell. 2013;49:1159–1166. doi: 10.1016/j.molcel.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 35.Atanassov BS, Koutelou E, Dent SY. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2011;585:2016–2023. doi: 10.1016/j.febslet.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 37.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 40.Xu H, Wang W, Li C, Yu H, Yang A, Wang B, Jin Y. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 2009;19:561–573. doi: 10.1038/cr.2009.31. [DOI] [PubMed] [Google Scholar]
- 41.Xu HM, et al. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J Biol Chem. 2004;279:23495–23503. doi: 10.1074/jbc.M400516200. [DOI] [PubMed] [Google Scholar]
- 42.Liao B, Jin Y. Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Res. 2010;20:332–344. doi: 10.1038/cr.2009.136. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- 46.Ramakrishna S, Suresh B, Lim KH, Cha BH, Lee SH, Kim KS, Baek KH. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011;20:1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- 47.Moretto-Zita M, Jin H, Shen Z, Zhao T, Briggs SP, Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci U S A. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yates A, Chambers I. The homeodomain protein Nanog and pluripotency in mouse embryonic stem cells. Biochem Soc Trans. 2005;33:1518–1521. doi: 10.1042/BST0331518. [DOI] [PubMed] [Google Scholar]
- 49.Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 50.Ye F, Zhou C, Cheng Q, Shen J, Chen H. Stem-cell-abundant proteins Nanog, Nucleostemin and Musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer. 2008;8:108. doi: 10.1186/1471-2407-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ, Zhou J, Ding YQ, Li JM. Overexpression of Nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;9:295–302. doi: 10.4161/cbt.9.4.10666. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Niu CS, Cheng CD. Pin1-Nanog expression in human glioma is correlated with advanced tumor progression. Oncol Rep. 2013;30:560–566. doi: 10.3892/or.2013.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgartel C, Schekman R, Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument ????, Bromage H, Tempst P, et al. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang L, Huang H, Zhou F, Schimmel J, Pardo CG, Zhang T, Barakat TS, Sheppard KA, Mickanin C, Porter JA, et al. RNF12 controls embryonic stem cell fate and morphogenesis in zebrafish embryos by targeting Smad7 for degradation. Mol Cell. 2012;46:650–661. doi: 10.1016/j.molcel.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 56.Hernebring M, Fredriksson A, Liljevald M, Cvijovic M, Norrman K, Wiseman J, Semb H, Nystrom T. Removal of damaged proteins during ES cell fate specification requires the proteasome activator PA28. Sci Rep. 2013;3:1381. doi: 10.1038/srep01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hernebring M, Brolen G, Aguilaniu H, Semb H, Nystrom T. Elimination of damaged proteins during differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:7700–7705. doi: 10.1073/pnas.0510944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- 60.Babaie Y, Herwig R, Greber B, Brink TC, Wruck W, Groth D, Lehrach H, Burdon T, Adjaye J. Analysis of Oct4-dependent transcriptional networks regulating self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2007;25:500–510. doi: 10.1634/stemcells.2006-0426. [DOI] [PubMed] [Google Scholar]
- 61.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- 63.Wilson A, Trumpp A. Bone-marrow haematopoietic-stem-cell niches. Nat Rev Immunol. 2006;6:93–106. doi: 10.1038/nri1779. [DOI] [PubMed] [Google Scholar]
- 64.Jude CD, Gaudet JJ, Speck NA, Ernst P. Leukemia and hematopoietic stem cells: balancing proliferation and quiescence. Cell Cycle. 2008;7:586–591. doi: 10.4161/cc.7.5.5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warr MR, Pietras EM, Passegue E. Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip Rev Syst Biol Med. 2011;3:681–701. doi: 10.1002/wsbm.145. [DOI] [PubMed] [Google Scholar]
- 66.Trumpp A, Essers M, Wilson A. Awakening dormant haematopoietic stem cells. Nat Rev Immunol. 2010;10:201–209. doi: 10.1038/nri2726. [DOI] [PubMed] [Google Scholar]
- 67.Lacombe J, Herblot S, Rojas-Sutterlin S, Haman A, Barakat S, Iscove NN, Sauvageau G, Hoang T. Scl regulates the quiescence and the long-term competence of hematopoietic stem cells. Blood. 2010;115:792–803. doi: 10.1182/blood-2009-01-201384. [DOI] [PubMed] [Google Scholar]
- 68.Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 70.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 71.Reavie L, Della Gatta G, Crusio K, Aranda-Orgilles B, Buckley SM, Thompson B, Lee E, Gao J, Bredemeyer AL, Helmink BA, et al. Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat Immunol. 2010;11:207–215. doi: 10.1038/ni.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pietras EM, Warr MR, Passegue E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195:709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsuoka S, Oike Y, Onoyama I, Iwama A, Arai F, Takubo K, Mashimo Y, Oguro H, Nitta E, Ito K, et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008;22:986–991. doi: 10.1101/gad.1621808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moran-Crusio K, Reavie LB, Aifantis I. Regulation of hematopoietic stem cell fate by the ubiquitin proteasome system. Trends Immunol. 2012;33:357–363. doi: 10.1016/j.it.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsumoto A, Nakayama KI. Role of key regulators of the cell cycle in maintenance of hematopoietic stem cells. Biochim Biophys Acta. 2013;1830:2335–2344. doi: 10.1016/j.bbagen.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 78.Li J. Quiescence regulators for hematopoietic stem cell. Exp Hematol. 2011;39:511–520. doi: 10.1016/j.exphem.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 79.Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J Biol Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- 80.Zeng S, Xu Z, Lipkowitz S, Longley JB. Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl) Blood. 2005;105:226–232. doi: 10.1182/blood-2004-05-1768. [DOI] [PubMed] [Google Scholar]
- 81.Goh EL, Zhu T, Leong WY, Lobie PE. c-Cbl is a negative regulator of GH-stimulated STAT5-mediated transcription. Endocrinology. 2002;143:3590–3603. doi: 10.1210/en.2002-220374. [DOI] [PubMed] [Google Scholar]
- 82.Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 83.Rathinam C, Thien CB, Langdon WY, Gu H, Flavell RA. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 2008;22:992–997. doi: 10.1101/gad.1651408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rathinam C, Thien CB, Flavell RA, Langdon WY. Myeloid leukemia development in c-Cbl RING finger mutant mice is dependent on FLT3 signaling. Cancer Cell. 2010;18:341–352. doi: 10.1016/j.ccr.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 85.Bacher U, Haferlach C, Schnittger S, Kohlmann A, Kern W, Haferlach T. Mutations of the TET2 and CBL genes: novel molecular markers in myeloid malignancies. Ann Hematol. 2010;89:643–652. doi: 10.1007/s00277-010-0920-6. [DOI] [PubMed] [Google Scholar]
- 86.Barresi V, Palumbo GA, Musso N, Consoli C, Capizzi C, Meli CR, Romano A, Di Raimondo F, Condorelli DF. Clonal selection of 11q CN-LOH and CBL gene mutation in a serially studied patient during MDS progression to AML. Leuk Res. 2010;34:1539–1542. doi: 10.1016/j.leukres.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 87.Fernandes MS, Reddy MM, Croteau NJ, Walz C, Weisbach H, Podar K, Band H, Carroll M, Reiter A, Larson RA, et al. Novel oncogenic mutations of CBL in human acute myeloid leukemia that activate growth and survival pathways depend on increased metabolism. J Biol Chem. 2010;285:32596–32605. doi: 10.1074/jbc.M110.106161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rocquain J, Carbuccia N, Trouplin V, Raynaud S, Murati A, Nezri M, Tadrist Z, Olschwang S, Vey N, Birnbaum D, et al. Combined mutations of ASXL1, CBL, FLT3, IDH1, IDH2, JAK2, KRAS, NPM1, NRAS, RUNX1, TET2 and WT1 genes in myelodysplastic syndromes and acute myeloid leukemias. BMC Cancer. 2010;10:401. doi: 10.1186/1471-2407-10-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rathinam C, Matesic LE, Flavell RA. The E3 ligase Itch is a negative regulator of the homeostasis and function of hematopoietic stem cells. Nat Immunol. 2011;12:399–407. doi: 10.1038/ni.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- 92.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, Hardwick J, Welcker M, Meijerink JP, Pieters R, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, Ferrando A, Aifantis I. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 95.Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- 96.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- 99.Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- 100.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 102.Thompson BJ, Jankovic V, Gao J, Buonamici S, Vest A, Lee JM, Zavadil J, Nimer SD, Aifantis I. Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J Exp Med. 2008;205:1395–1408. doi: 10.1084/jem.20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Onoyama I, Tsunematsu R, Matsumoto A, Kimura T, de Alboran IM, Nakayama K, Nakayama KI. Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J Exp Med. 2007;204:2875–2888. doi: 10.1084/jem.20062299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nakayama K, Nagahama H, Minamishima YA, Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T, Nakayama KI. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev Cell. 2004;6:661–672. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- 105.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodriguez S, Wang L, Mumaw C, Srour EF, Lo Celso C, Nakayama K, Carlesso N. The SKP2 E3 ligase regulates basal homeostasis and stress-induced regeneration of HSCs. Blood. 2011;117:6509–6519. doi: 10.1182/blood-2010-11-321521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J, Han F, Wu J, Lee SW, Chan CH, Wu CY, Yang WL, Gao Y, Zhang X, Jeong YS, et al. The role of Skp2 in hematopoietic stem cell quiescence, pool size, and self-renewal. Blood. 2011;118:5429–5438. doi: 10.1182/blood-2010-10-312785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci U S A. 2007;104:5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, et al. Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell. 2010;7:391–402. doi: 10.1016/j.stem.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 110.Abbas HA, Maccio DR, Coskun S, Jackson JG, Hazen AL, Sills TM, You MJ, Hirschi KK, Lozano G. Mdm2 is required for survival of hematopoietic stem cells/progenitors via dampening of ROS-induced p53 activity. Cell Stem Cell. 2010;7:606–617. doi: 10.1016/j.stem.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 112.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 113.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 114.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 115.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16:314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Parmar K, Kim J, Sykes SM, Shimamura A, Stuckert P, Zhu K, Hamilton A, Deloach MK, Kutok JL, Akashi K, et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells. 2010;28:1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–457. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 118.Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, et al. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- 119.Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, Cotsarelis G. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature. 2007;447:316–320. doi: 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- 120.Solanas G, Benitah SA. Regenerating the skin: a task for the heterogeneous stem cell pool and surrounding niche. Nat Rev Mol Cell Biol. 2013;14:737–748. doi: 10.1038/nrm3675. [DOI] [PubMed] [Google Scholar]
- 121.Perez-Losada J, Balmain A. Stem-cell hierarchy in skin cancer. Nat Rev Cancer. 2003;3:434–443. doi: 10.1038/nrc1095. [DOI] [PubMed] [Google Scholar]
- 122.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 123.Silva-Vargas V, Lo Celso C, Giangreco A, Ofstad T, Prowse DM, Braun KM, Watt FM. Beta-catenin and Hedgehog signal strength can specify number and location of hair follicles in adult epidermis without recruitment of bulge stem cells. Dev Cell. 2005;9:121–131. doi: 10.1016/j.devcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 124.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 125.Han G, Li AG, Liang YY, Owens P, He W, Lu S, Yoshimatsu Y, Wang D, Ten Dijke P, Lin X, et al. Smad7-induced beta-catenin degradation alters epidermal appendage development. Dev Cell. 2006;11:301–312. doi: 10.1016/j.devcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 126.Perry WL, Hustad CM, Swing DA, O'Sullivan TN, Jenkins NA, Copeland NG. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet. 1998;18:143–146. doi: 10.1038/ng0298-143. [DOI] [PubMed] [Google Scholar]
- 127.Rossi M, Aqeilan RI, Neale M, Candi E, Salomoni P, Knight RA, Croce CM, Melino G. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci U S A. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rossi M, De Simone M, Pollice A, Santoro R, La Mantia G, Guerrini L, Calabro V. Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle. 2006;5:1816–1822. doi: 10.4161/cc.5.16.2861. [DOI] [PubMed] [Google Scholar]
- 129.Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- 130.Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 131.Fang D, Elly C, Gao B, Fang N, Altman Y, Joazeiro C, Hunter T, Copeland N, Jenkins N, Liu YC. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat Immunol. 2002;3:281–287. doi: 10.1038/ni763. [DOI] [PubMed] [Google Scholar]
- 132.Melino G, Gallagher E, Aqeilan RI, Knight R, Peschiaroli A, Rossi M, Scialpi F, Malatesta M, Zocchi L, Browne G, et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ. 2008;15:1103–1112. doi: 10.1038/cdd.2008.60. [DOI] [PubMed] [Google Scholar]
- 133.Candi E, Rufini A, Terrinoni A, Dinsdale D, Ranalli M, Paradisi A, De Laurenzi V, Spagnoli LG, Catani MV, Ramadan S, et al. Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ. 2006;13:1037–1047. doi: 10.1038/sj.cdd.4401926. [DOI] [PubMed] [Google Scholar]
- 134.Galli F, Rossi M, D'Alessandra Y, De Simone M, Lopardo T, Haupt Y, Alsheich-Bartok O, Anzi S, Shaulian E, Calabro V, et al. MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J Cell Sci. 2010;123:2423–2433. doi: 10.1242/jcs.061010. [DOI] [PubMed] [Google Scholar]
- 135.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 136.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 137.Hart M, Concordet JP, Lassot I, Albert I, del los Santos R, Durand H, Perret C, Rubinfeld B, Margottin F, Benarous R, et al. The F-box protein beta-TrCP associates with phosphorylated beta-catenin and regulates its activity in the cell. Curr Biol. 1999;9:207–210. doi: 10.1016/s0960-9822(99)80091-8. [DOI] [PubMed] [Google Scholar]
- 138.Ilyas M, Tomlinson IP, Rowan A, Pignatelli M, Bodmer WF. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc Natl Acad Sci U S A. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 140.Kudo Y, Guardavaccaro D, Santamaria PG, Koyama-Nasu R, Latres E, Bronson R, Yamasaki L, Pagano M. Role of F-box protein betaTrcp1 in mammary gland development and tumorigenesis. Mol Cell Biol. 2004;24:8184–8194. doi: 10.1128/MCB.24.18.8184-8194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Belaidouni N, Peuchmaur M, Perret C, Florentin A, Benarous R, Besnard-Guerin C. Overexpression of human beta TrCP1 deleted of its F box induces tumorigenesis in transgenic mice. Oncogene. 2005;24:2271–2276. doi: 10.1038/sj.onc.1208418. [DOI] [PubMed] [Google Scholar]
- 142.Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 143.Babaei-Jadidi R, Li N, Saadeddin A, Spencer-Dene B, Jandke A, Muhammad B, Ibrahim EE, Muraleedharan R, Abuzinadah M, Davis H, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208:295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 145.Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 146.Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- 147.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 148.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chu J, Hong NA, Masuda CA, Jenkins BV, Nelms KA, Goodnow CC, Glynne RJ, Wu H, Masliah E, Joazeiro CA, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci U S A. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 152.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 154.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 155.Cao J, Yan Q. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front Oncol. 2012;2:26. doi: 10.3389/fonc.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 157.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 160.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 161.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Laribee RN, Fuchs SM, Strahl BD. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Genes Dev. 2007;21:737–743. doi: 10.1101/gad.1541507. [DOI] [PubMed] [Google Scholar]
- 165.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–388. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, Konig A, Snaidero N, Vogel T, Shchebet A, Begus-Nahrmann Y, et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 167.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, Golomb L, Pribluda A, Zhang F, Haj-Yahya M, et al. RNF20 and USP44 regulate stem cell differentiation by modulating H2B monoubiquitylation. Mol Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Vethantham V, Yang Y, Bowman C, Asp P, Lee JH, Skalnik DG, Dynlacht BD. Dynamic loss of H2B ubiquitylation without corresponding changes in H3K4 trimethylation during myogenic differentiation. Mol Cell Biol. 2012;32:1044–1055. doi: 10.1128/MCB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics. 2010;5:460–468. doi: 10.4161/epi.5.6.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 171.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Piao S, Liu Y, Hu J, Guo F, Ma J, Sun Y, Zhang B. USP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinoma. PLoS One. 2012;7:e42540. doi: 10.1371/journal.pone.0042540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, Pang D. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J Cancer Res Clin Oncol. 2011;137:1245–1253. doi: 10.1007/s00432-011-0998-9. [DOI] [PubMed] [Google Scholar]
- 174.Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–251. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 176.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 177.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 178.Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- 179.Wei J, Zhai L, Xu J, Wang H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J Biol Chem. 2006;281:22537–22544. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- 180.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chen A, Kleiman FE, Manley JL, Ouchi T, Pan ZQ. Autoubiquitination of the BRCA1*BARD1 RING ubiquitin ligase. J Biol Chem. 2002;277:22085–22092. doi: 10.1074/jbc.M201252200. [DOI] [PubMed] [Google Scholar]
- 186.Xia Y, Pao GM, Chen HW, Verma IM, Hunter T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem. 2003;278:5255–5263. doi: 10.1074/jbc.M204591200. [DOI] [PubMed] [Google Scholar]
- 187.Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477:179–184. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 190.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 191.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave LM, et al. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med. 2013;210:2641–2659. doi: 10.1084/jem.20131141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 196.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 197.Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell. 2006;22:383–394. doi: 10.1016/j.molcel.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 198.Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 199.Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502:249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- 200.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 201.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 202.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 205.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 206.Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, Spruck C, Grander D, Lendahl U, Sangfelt O. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- 207.Lee JW, Soung YH, Kim HJ, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. Mutational analysis of the hCDC4 gene in gastric carcinomas. Eur J Cancer. 2006;42:2369–2373. doi: 10.1016/j.ejca.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 208.Low J, Blosser W, Dowless M, Ricci-Vitiani L, Pallini R, de Maria R, Stancato L. Knockdown of ubiquitin ligases in glioblastoma cancer stem cells leads to cell death and differentiation. J Biomol Screen. 2012;17:152–162. doi: 10.1177/1087057111422565. [DOI] [PubMed] [Google Scholar]
- 209.Micel LN, Tentler JJ, Smith PG, Eckhardt GS. Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J Clin Oncol. 2013;31:1231–1238. doi: 10.1200/JCO.2012.44.0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chan CH, Morrow JK, Li CF, Gao Y, Jin G, Moten A, Stagg LJ, Ladbury JE, Cai Z, Xu D, et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell. 2013;154:556–568. doi: 10.1016/j.cell.2013.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Chanet S, Schweisguth F. Regulation of epithelial polarity by the E3 ubiquitin ligase Neuralized and the Bearded inhibitors in Drosophila. Nat Cell Biol. 2012;14:467–476. doi: 10.1038/ncb2481. [DOI] [PubMed] [Google Scholar]
- 212.Benhra N, Vignaux F, Dussert A, Schweisguth F, Le Borgne R. Neuralized promotes basal to apical transcytosis of delta in epithelial cells. Mol Biol Cell. 2010;21:2078–2086. doi: 10.1091/mbc.E09-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tenenhaus C, Subramaniam K, Dunn MA, Seydoux G. PIE-1 is a bifunctional protein that regulates maternal and zygotic gene expression in the embryonic germ line of Caenorhabditis elegans. Genes Dev. 2001;15:1031–1040. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Tenlen JR, Molk JN, London N, Page BD, Priess JR. MEX-5 asymmetry in one-cell C. elegans embryos requires PAR-4- and PAR-1-dependent phosphorylation. Development. 2008;135:3665–3675. doi: 10.1242/dev.027060. [DOI] [PubMed] [Google Scholar]
- 215.Li S, Wang C, Sandanaraj E, Aw SS, Koe CT, Wong JJ, Yu F, Ang BT, Tang C, Wang H. The SCFSlimb E3 ligase complex regulates asymmetric division to inhibit neuroblast overgrowth. EMBO Rep. 2014;15:165–174. doi: 10.1002/embr.201337966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Anon. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Morais-de-Sá E, Vega-Rioja A, Trovisco V, St Johnston D. Oskar is targeted for degradation by the sequential action of Par-1, GSK-3, and the SCF-slimb ubiquitin ligase. Dev Cell. 2013;26:303–314. doi: 10.1016/j.devcel.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Vertegaal AC. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 220.Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 221.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kim TW, Guan S, Burlingame AL, Wang ZY. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol Cell. 2011;43:561–571. doi: 10.1016/j.molcel.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 224.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]