Figure 2.
PIH-N Domain Structure and Tel2 Interactions
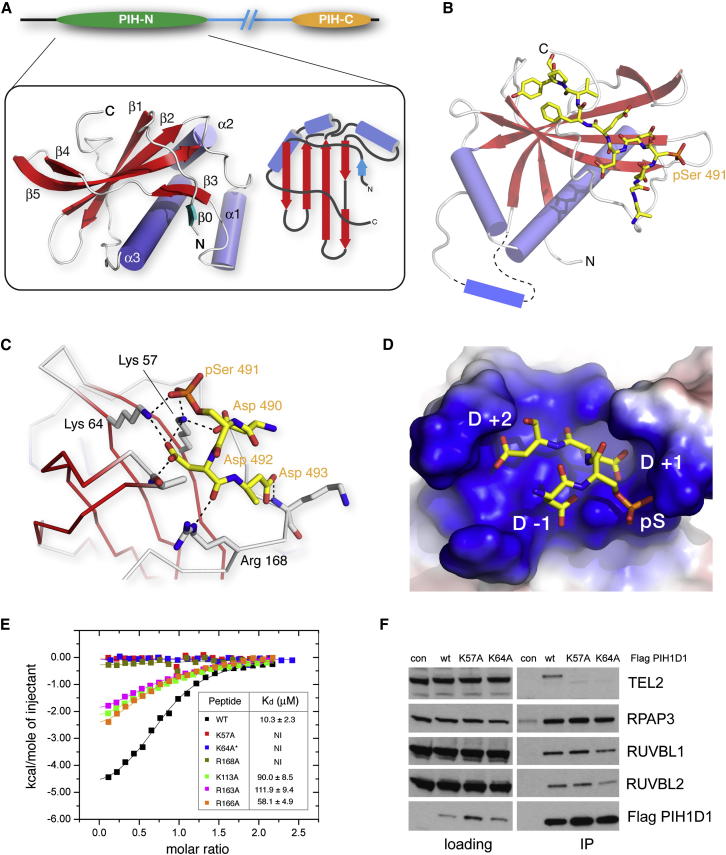
(A) Left, ribbons representation of the uncomplexed PIH1D1 PIH-N domain. Right, schematic representation of the overall α + β topology. β0 (cyan) contains four residues of vector encoded sequence that most likely mimics the conformation of native sequence that was removed in construction of the crystallizable fragment.
(B) Structure of the PIH-N domain of PIH1D1 bound to a TEL2 phosphopeptide. The protein is shown as ribbons and the phosphopeptide as a stick representation. The loop containing α1 is disordered in the phosphopeptide complex structure and is shown schematically.
(C) The core DpSDD motif in TEL2 is secured via a network of salt bridge and hydrogen bonding interactions with three basic residues: Lys57, Lys64, and Arg168.
(D) Aspartates in the pSer +1 and +2 positions (and, to a lesser extent the −1 position) within the TEL2 peptide pack tightly against the protein surface.
(E) ITC isotherms show that mutation of the core-motif-interacting side chains from Lys57, Lys64, and Arg168 are most deleterious to TEL2 binding. The asterisk shows that the titration with K64A was carried out at higher concentration/molar ratio, but, nonetheless, no interaction was detectable.
(F) Lys57 and Lys64, but not R2TP, are essential for TEL2 binding components in vivo. RUVBL1, RUVBL2, RPAP3, and TEL2 were immunoprecipitated from HEK293T cells transiently transfected with FLAG-tagged WT PIH1D1 or PIH1D1 with K64A and K57A mutations.