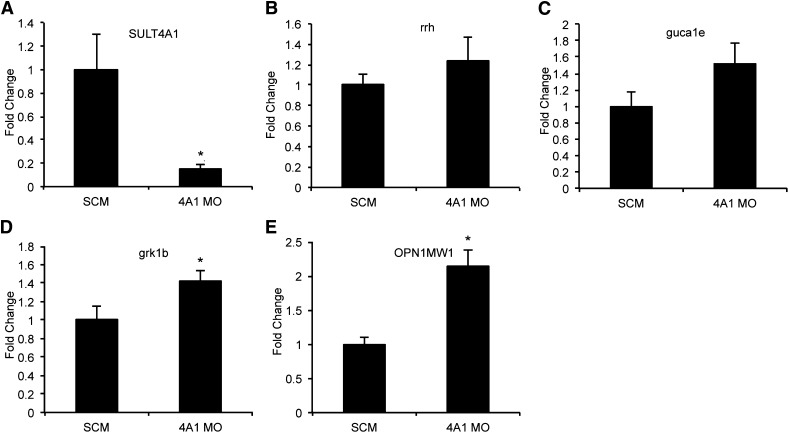
Fig. 3.
qPCR verification of differentially expressed phototransduction genes observed in RNA-seq data at 72 hpf. SuperScript III was used to generate cDNA from total RNA. Message level was determined on a 7900HT Sequence Detection System using predesigned TaqMan gene expression assays. Values represent average relative mRNA expression (n = 3) +/− the standard error of the mean. (A) SULT4A1: SCM (1.00 +/− 0.31); SULT4A1 MO (0.15 +/− 0.04); P = 0.0253. (B) rrh: SCM (1.00 +/− 0.12); SULT4A1 MO (1.24 +/− 0.23); P = 0.1917. (C) guca1e: SCM (1.00 +/− 0.17); SULT4A1 MO (1.52 +/− 0.26); P = 0.0822. (D) grk1b: SCM (1.00 +/− 0.14); SULT4A1 MO (1.43 +/− 0.11); P = 0.0392. (E) OPN1MW1: SCM (1.00 +/− 0.10); SULT4A1 MO (2.15 +/− 0.22); P = 0.0047. *P < 0.05 as compared with SCM-injected embryos.