The use of advanced next-generation sequencing techniques, including global run-on sequencing 1, has captured multiple antisense and sense non-coding (nc) RNA transcripts produced bidirectionally around promoters. Other ncRNAs, such as enhancer RNAs and lncRNAs, are transcribed from a large fraction of genome 2, 3. Together, these insights and others make clear that the complexity of the dynamics and landscape of the regulation of active genes is staggering.
The promoters of some actively transcribed genes are likely to be embedded in a “cloud” of sense and antisense ncRNAs, the mass balance of which could modify gene expression in a fashion similar to the way in which the mass action law determines the direction of chemical reactions involving e.g. spacial competition, steric hindrance, active RNA-duplex-mediated mechanisms and the epigenetic regulation of transcription 3, 4. Regulation of (productive) transcription based on these principles could help to understand why gene expression and physiological effects are so consistent in millions of cells in given tissues despite the inevitable small errors and mismatches that take place in individual cells.
In support of the ‘ncRNA cloud concept’ at active promoters (Fig 1), we 4 and others 5 have found that small hairpin (sh) RNAs that bind to specific promoter areas of endogenous genes without any sequence homology with mRNA—i.e. they do not act through classical RNAi—can cause significant up- or down-regulation of target genes in vitro and in vivo. We also found that small hairpin structures are required for the activity and that the mechanism involves epigenetic changes such as histone methylation and acetylation 4. Cellular factors such as AGO2, LSD1 and CBP/p300 have been suggested as mediators of these effects. Given that epigenetic changes are reversible, the therapeutic potential of this approach is obvious 6.
Figure 1.
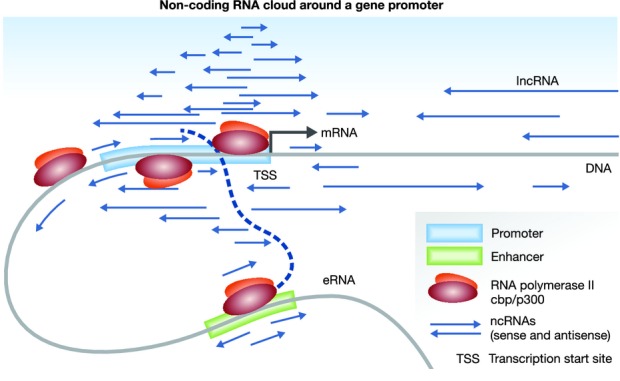
“A cloud concept” of non-coding RNAs around promoters.
The promoters of some actively transcribed genes are surrounded by multiple non-coding sense and antisense RNA transcripts and their mass balance could modify gene expression in a similar fashion as the mass action law determines the direction of chemical reactions.
The clinical potential of siRNAs, miRNAs and ncRNAs has been widely recognized. Antagomirs, antagoNATs (NAT stands for natural antisense transcripts), siRNAs and antisense oligos are currently being tested for various applications 7. By following the principles of the mass action law in chemistry and providing excess exogenous shRNAs that specifically bind to the promoter or to the promoter-associated sense or antisense ncRNAs, we should be able to modify the mass balance of both RNA molecules and mechanisms such as spacial competition, steric hindrance and recruitment of transcription regulators to the promoter, thus resulting in changes in coding gene transcription.
This approach is likely to be of value for applications that require the upregulation of endogenous genes from their genomic loci, including genes where several natural isoforms are produced as a result of the natural stimulus 4. The majority of current drugs and genetically active tools are based on inhibitory mechanisms, whereas there is a shortage of agents capable of increasing the activity of mediators or pathways, such as growth factors and tumor suppressor genes. Short shRNAs complementary to the target gene promoter region could also improve specificity of the intended therapeutic effects.
While we can expect further discoveries about the overall role of ncRNAs in regulating gene expression programs, the direct utilization of shRNAs and principles of the mass action may simplify the concept and bring us one step closer to clinical applications.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Core LJ, Waterfall JJ, Lis JT. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, et al. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Lam MT, Glass CK. Cardiovasc Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen MP, et al. Circ Res. 2009;105:604–609. doi: 10.1161/CIRCRESAHA.109.200774. [DOI] [PubMed] [Google Scholar]
- Morris K, et al. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husso T, et al. Biomol Concepts. 2009;2:127–134. doi: 10.1515/bmc.2011.012. [DOI] [PubMed] [Google Scholar]
- Wahlestedt C. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]


