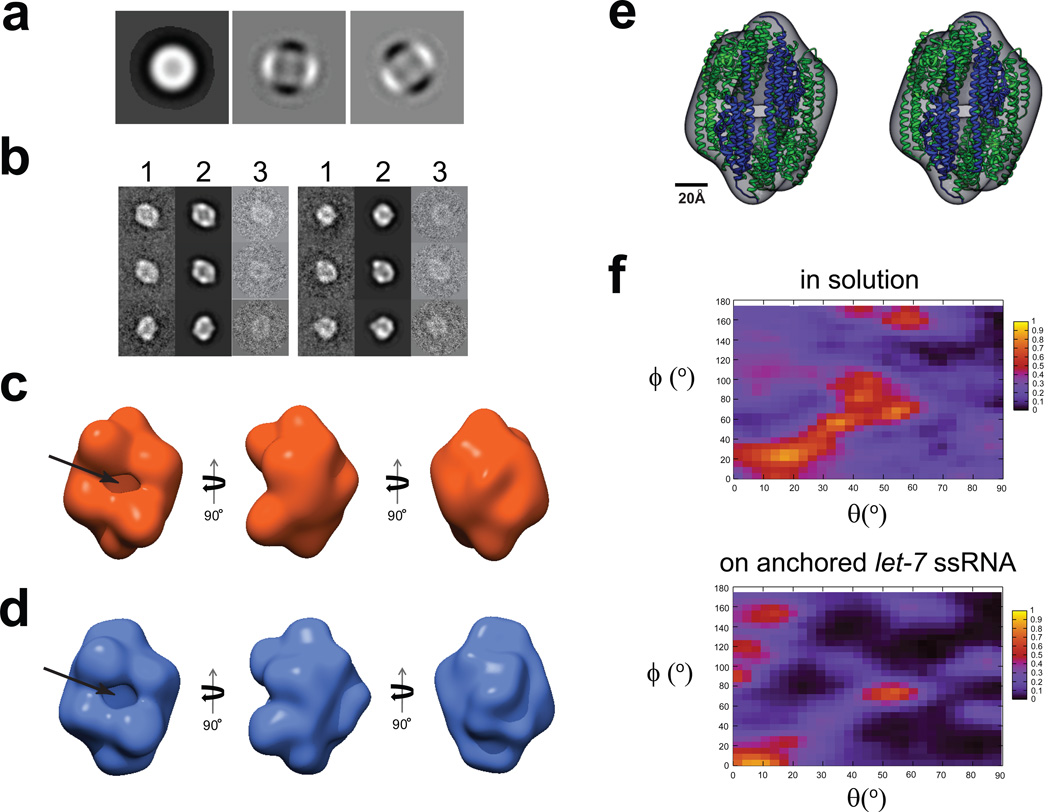
Figure 6. 3D reconstructions of C3PO assembled on ssRNA-ChemiC films.
(a) First three eigenimages after the multivariate statistical analysis of the center-aligned raw dataset. Strong 2-fold symmetry is evident from the second and third eigenimages, which were also seen in the dataset of the C3PO/DNA complexes in solution (Figure S4a).
(b) Representative class sums (1), projections from the 3D reconstruction (2) at the corresponding orientations, and the corresponding raw data (3) agree well with each other, and clearly reveal a football-shaped structure with a hollow interior. The matching among these three sets suggests that the 3D reconstruction does reflect the features in the raw images of the C3PO/ssRNA complexes on the ssRNA-ChemiC films.
(c) Three different views of the 3D reconstruction of C3PO/anchored let-7 ssRNA complexes. The side port is ~15 × 25 Å2. The volume is surfaced to enclose a mass of ~240 kDa, the expected mass of a C3PO octamer. If we threshold the density to reflect a density of a hexamer (180 kDa), the densities started to show disconnections, suggesting the 3D map likely represents an octamer.
(d) Three views of the 3D reconstruction calculated from negatively stained C3PO/ssDNA complexes in solution. This control reconstruction was obtained completely independently of the map in (c). It shows very similar features. The size of the side port is almost the same.
(e) Stereo view of the X-ray structure of the human C3PO octamer (ribbons; PDB code: 3PJA; blue for TRAX and green for translin subunits) docked into the 3D model (semi-transparent surface map in grey) of C3PO on the ssRNA-ChemiC-coated grids. The octameric structural model was positioned such that the two TRAX subunits are at the side port. The crystal structure shows a small gap at the TRAX dimer interface, which was proposed to become wider in order to admit the ssRNA substrates.
(f) Heat maps showing the orientational distribution for individual C3PO particles bound with ssDNA in solution (top) and that for C3PO complexes assembled on ssRNA-ChemiC films (bottom). The angles θand ϕfollow the standard definition in SPIDER.