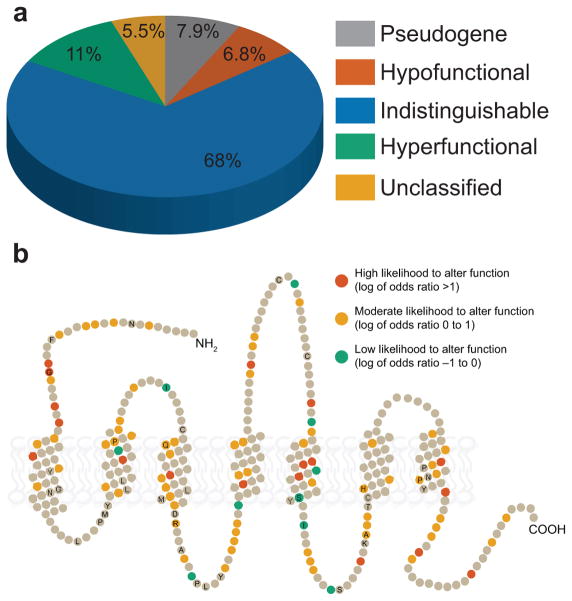
Figure 4.
Summary of functional variation. (a) The type of functional differences among 27 odorant receptors of 1092 participants from the 1000 Genomes Project. Note that pseudogenes account for a small portion of the variability relative to missense variations. (b) Snake plot of a typical odorant receptor showing residues where SNPs alter the function of the receptor. Amino acid residues that did not vary between any of the minor alleles and their reference allele are shown in gray. The remaining residues are colored according to the odds that they alter function given our current dose-response data. Amino acid positions conserved in at least 90% of the receptors are labeled with their single-letter amino acid code.