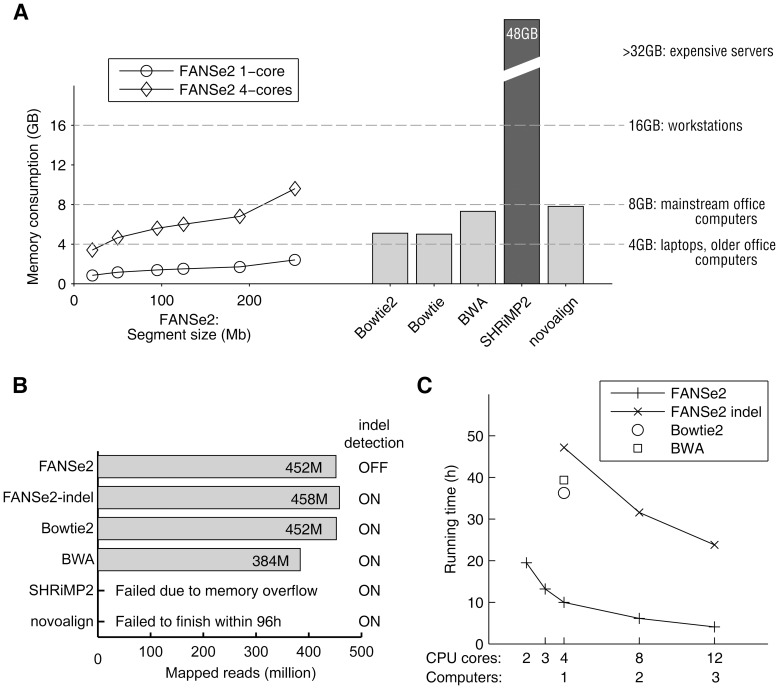
Figure 3. Scalability, sensitivity and speed of FANSe2 compared to other algorithms.
(A) Memory consumption of the tested algorithms when mapping 75-nt reads to human genome. The memory consumption of FANSe2 using 1 CPU core and 4 CPU cores are indicated using circles and diamonds, respectively. SHRiMP2 failed to run this test in our 16 GB memory system; thus its memory consumption was taken from its manual. (B, C) Mapping data from an entire Illumina HiSeq-2000 flowcell (608 M 75-nt reads) to masked human genome. (B) The number of reads mapped by the tested algorithms using one computer (4 CPU cores). FANSe2 was tested with indel detection on and off, respectively. SHRiMP2 failed to run in our system due to its high memory consumption. Novoalign failed to finish the task within 96 hours. (C) The time to perform this mapping using different number of CPU cores and computers. Plus sign: FANSe2 without indel detection; cross: FANSe2 with indel detection. Bowtie2 (circle) and BWA (rectangle) do not support automatic parallelization across multiple computers.