Table 1.
A summary of epigenetic marks, their types (wherever applicable), and the NGS-based assays used to map their location and distribution
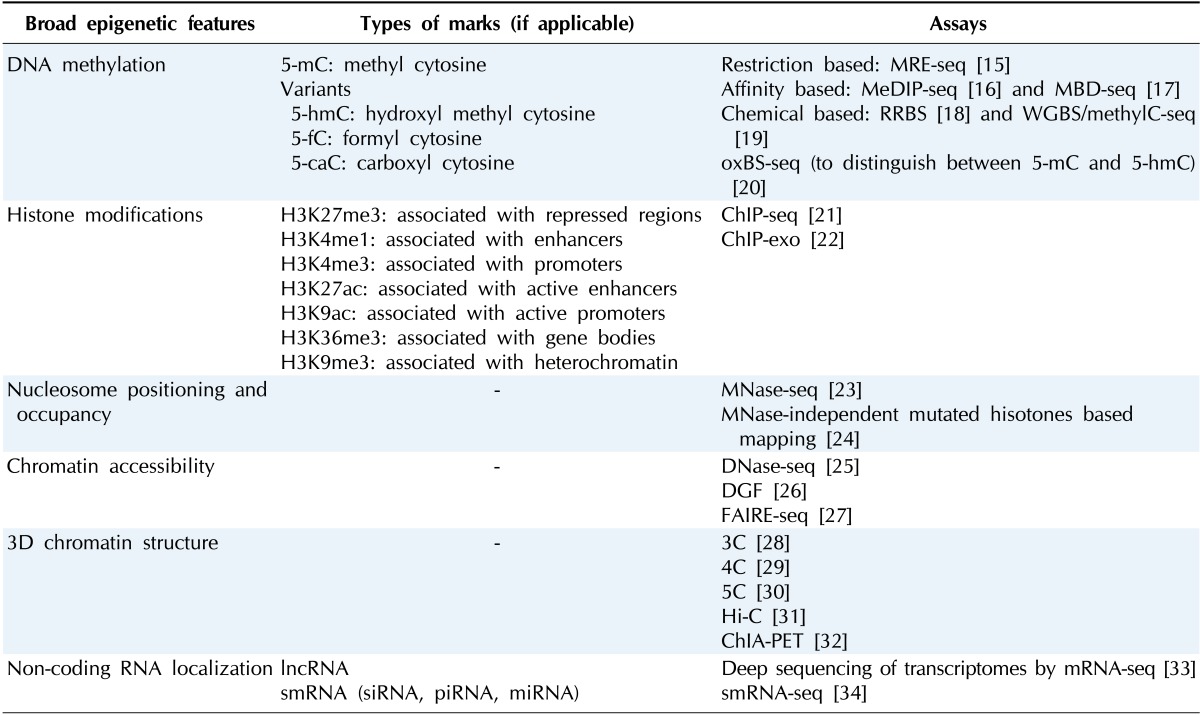
MRE-seq, methylation-sensitive restriction enzyme sequencing; MeDIP-seq, methylated DNA immunoprecipitation sequencing; MBD-seq, methyl-CpG-binding domain protein sequencing; RRBS, reduced representation bisulfite sequencing; WGBS, whole-genome bisulfite sequencing; oxBS-seq, oxidative bisulfite sequencing; ChIP-seq, chromatin immunoprecipitation sequencing; ChIP-exo, chromatin immunoprecipitation-exonuclease; DGF, digital genomic footprint; FAIRE-seq, formaldehyde-assisted isolation of regulatory elements sequencing; ChIA-PET, chromatin interaction analysis by paired-end tag sequencing.
