Figure 1.
Jmjd4 Interacts with the eRF1/eRF3a Translational Termination Complex in an Activity-Dependent Manner
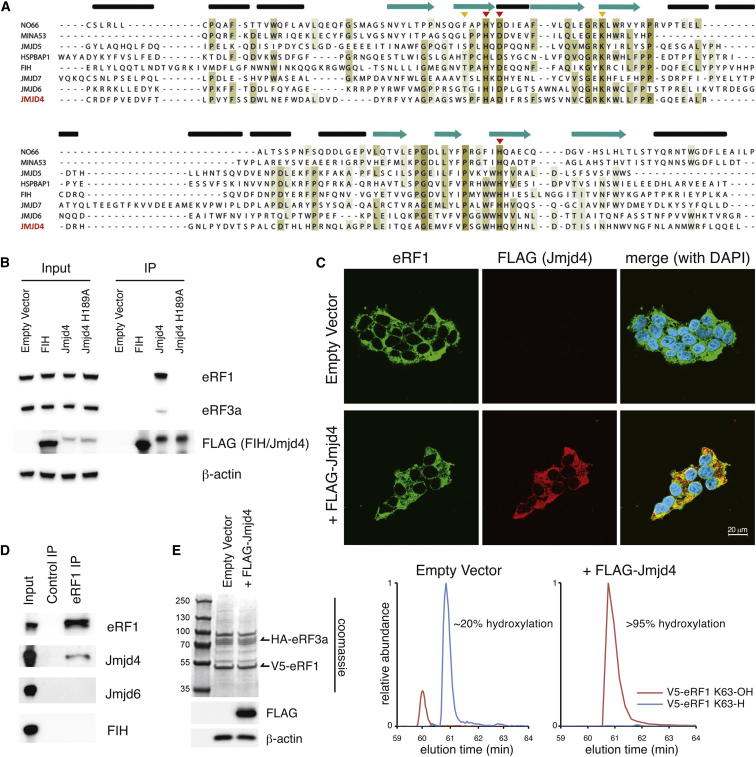
(A) Protein sequences of human JmjC domains were aligned and shaded using Jalview. The Jmjd6 secondary structure, as defined by crystallographic analysis (Mantri et al., 2010), is indicated with α helices (cylinders) and β strands (arrows). The conserved double-stranded β helix core is in cyan (arrows). Triangles indicate residues binding Fe(II) (red) and 2OG (yellow). Mutation of the first Fe(II)-binding residue (His189 in Jmjd4) is predicted to inhibit activity.
(B) Anti-FLAG immunoprecipitates of cell extracts from the indicated HEK293T cell lines were immunoblotted for endogenous eRF1 and eRF3a. Input (5%) = cell extract prior to immunoprecipitation (IP). eRF1 levels were quantified by densitometry analysis using NIH ImageJ.
(C) HEK293T cell lines were immunostained for eRF1 (green) and FLAG-Jmjd4 (red). Nuclei were visualized with DAPI (blue).
(D) Endogenous eRF1 and Jmjd4 interact. eRF1 was immunoprecipitated from HEK293T extracts prior to immunoblot for the indicated proteins.
(E) Overexpressed Jmjd4 promotes hydroxylation of overexpressed eRF1 at K63. Left: Coomassie gel showing 5% input following anti-V5 purification of the V5-eRF1/HA-eRF3a complex from HEK293T cells overexpressing empty vector or FLAG-Jmjd4 (immunoblot bottom panel). The remainder of the sample (95%) was digested with Arg-C in-solution prior to LC-MS analyses. The chromatograms indicate the elution time and relative abundance of extracted ion masses corresponding to unhydroxylated (blue) and K63-hydroxylated (red) eRF148–65 ([M+H]3+; K63-H: m/z 646.68; K63-OH: m/z 652.00) in the absence (middle) and presence (right) of FLAG-Jmjd4. See also Figure S1.