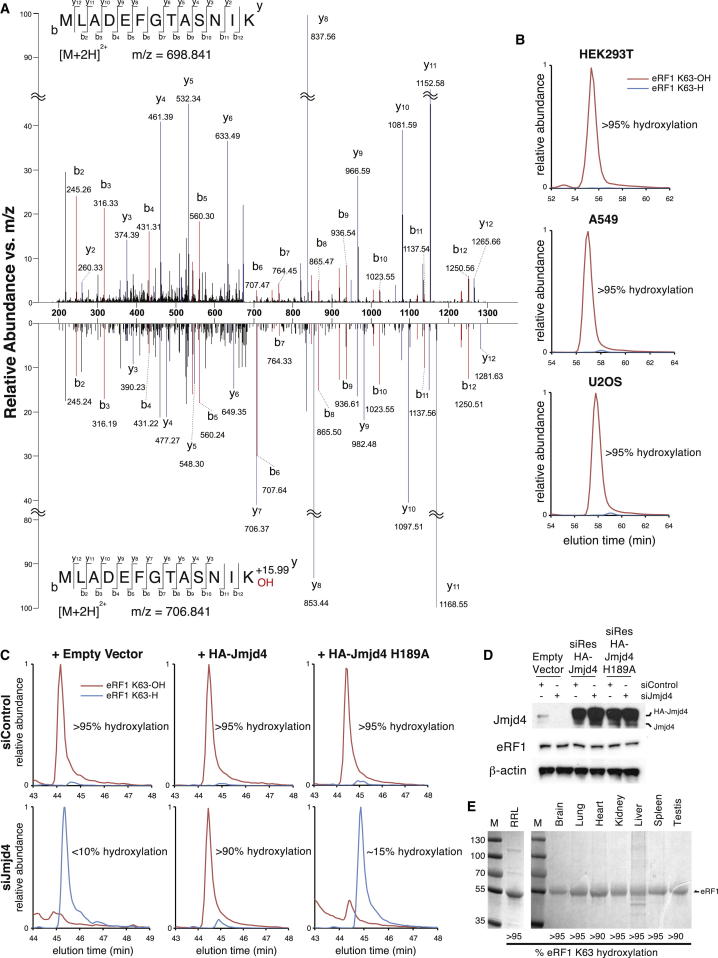
Figure 3.
Endogenous eRF1 K63 Hydroxylation Is Abundant, Ubiquitous, and Dependent on Jmjd4 Activity
(A) HEK293T eRF1 was trypsinized prior to MS/MS. Spectra show a +16 Da mass shift in all detected y ion fragments (y2–y12) (blue) when comparing the upper (unhydroxylated) and lower (hydroxylated) panels. In contrast, the masses of detected b ion fragments (b2–b12) (red) are consistent with their predicted values, indicating K63 hydroxylation. This was confirmed with Arg-C proteolysis (Figure S3A).
(B) LC-MS analyses of trypsinized eRF1 indicate abundant K63 hydroxylation in multiple cell types. EICs show relative abundance of unhydroxylated (blue) versus K63 hydroxylated (red) eRF151–63 ([M+H]2+; K63-H: m/z 698.842; K63-OH: m/z 706.840).
(C) EICs demonstrate that eRF1 hydroxylation is dependent on Jmjd4 activity ([M+H]2+; K63-H: m/z 701.852; K63-OH: m/z 709.850). The masses are +3 Da relative to (B) due to SILAC (stable isotope labeling by/with amino acids in cell culture) with K+6. HeLa cells expressing empty vector (left), siRNA-resistant HA-Jmjd4 (middle), or siRNA-resistant HA-Jmjd4 H189A mRNAs (right) were transfected with control (top row) or Jmjd4 siRNA (bottom row) prior to SILAC labeling and LC-MS quantitation of hydroxylation in newly synthesized eRF1. Note that although hydroxylation of newly synthesized eRF1 is <15% following Jmjd4 siRNA, total eRF1 is ∼50% hydroxylated due to persisting eRF1 synthesized prior to siRNA (data not shown).
(D) Cell extracts from (C) were immunoblotted for the indicated proteins. eRF1 levels were quantified by densitometry analysis.
(E) Similar analyses in rodent tissues indicate that eRF1 hydroxylation is physiologically relevant and conserved. Rabbit reticulocyte lysate (RRL) and the indicated mouse tissues were diluted or homogenized in lysis buffer prior to eRF1 purification, trypsinolysis, and MS analyses. See also Figure S3.