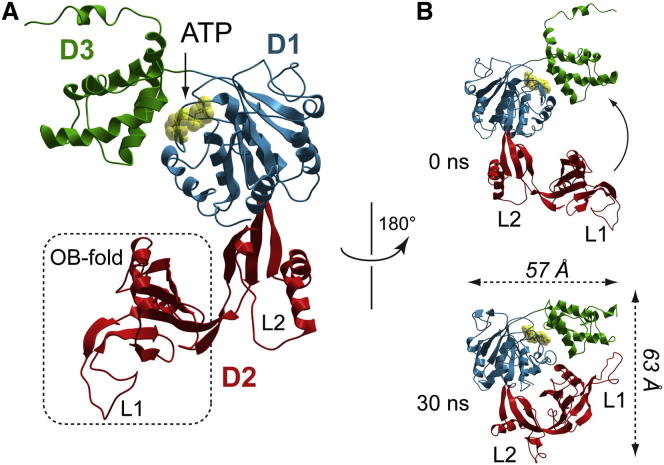
Figure 1.
All-Atom Model of the M. kandleri TIP49 Protein
(A) Domain organization of mkTIP49 monomers. The AAA+ D1 core is colored in blue; the D2 insertion domain, composed of the OB-fold (dotted rectangle) and L2, is colored in red; and the D3 regulatory domain is colored in green. ATP (yellow) is located in the catalytic pocket, formed at the interface between D1 and D3. Note the high level of sequence similarity between mkTIP49 and its human paralogs: 45% of identity with hsTIP49a and 42% with hsTIP49b.
(B) Major conformational transitions of mkTIP49 monomers in the course of 30 ns MD simulations are associated with OB-fold flexibility (arrow). Graphics were produced using the ICM-Pro software package. See also Figure S1 for sequence alignments between mkTIP49 and its eukaryotic paralogs TIP49a and TIP49b.