Figure 6. DNMT1 demonstrates dynamic interaction with Kir4.1 DNA during development.
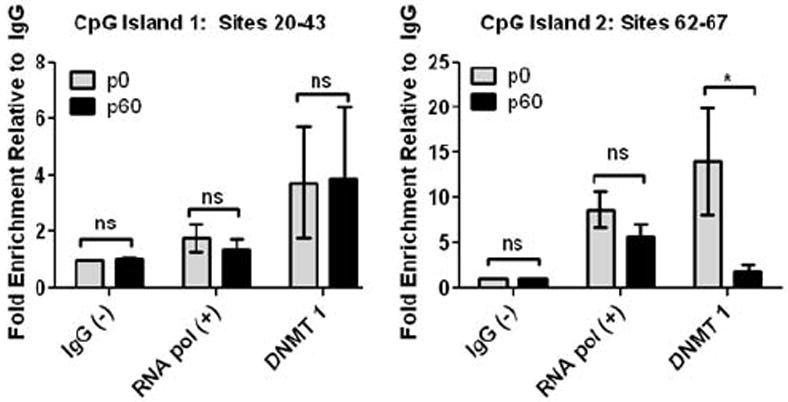
Chromatin immunoprecipitation and subsequent qPCR analysis (ChIP-qPCR) was used to assess DNMT1 interaction with Kir4.1 at two regions that demonstrated contrasting levels and age-related changes in methylation. IgG was used as a negative control and for normalization of background signal. RNA polymerase II was used as a positive control. Sites 20-43 found in CpG Island 1 contained low, stagnant levels of methylation during aging, while sites 62-67 found in CpG Island 2 contained higher levels of methylation that decreased significantly during aging. (A) ChIP analysis reveals that sites assessed in CpG island 1 demonstrate little change in DNMT1 interaction during aging from p0 to p60, fold enrichment of 3.74 and 3.89, respectively. (B) Conversely, sites assessed in CpG island 2 exhibit a decrease in DNMT 1 interaction during development, fold enrichment of 14.02 at p0 versus 1.81 at p60. For each age n=3; error bars represent s.e.m. * (P<0.05).
